Read Sentinel-1 RTC data processed by ASF using xr.open_mfdataset()#
This notebook demonstrates working with Sentinel-1 RTC imagery that has been processed on the ASF On-Demand server and downloaded locally.
The access point for data in this notebook is a directory containing un-zipped directories of RTC scenes.
This notebook will detail how to use the xarray function xr.open_mfdataset(). While this approach works and is very useful for working with large stacks of data with associated metadata, you will also see an example of the limitations of this approach due to certain characteristics of the dataset. You can read more about xr.open_mfdataset() here.
Learning goals:
Organize large set of geotiff files stored locally
Read data into memory using Xarray’s
open_mfdataset()method.
Software and setup#
Show code cell source
import dask
from dask.distributed import Client
from functools import partial
import geopandas as gpd
import matplotlib.pyplot as plt
import numpy as np
import os
import pandas as pd
import pathlib
import re
import xarray as xr
Initialize a dask.distributed client:
Note
On my local machine, I ran into issues initially when the cluster was running on processes rather than threads. This caused system memory issues and many dead kernels. Setting processes=False seemed to fix these issues
client = Client(processes=False)
client
Client
Client-9e4d56f7-0ead-11f0-88cb-c858b3e3e5bd
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: http://192.168.86.116:8787/status |
Cluster Info
LocalCluster
0f24e456
| Dashboard: http://192.168.86.116:8787/status | Workers: 1 |
| Total threads: 14 | Total memory: 38.76 GiB |
| Status: running | Using processes: False |
Scheduler Info
Scheduler
Scheduler-1a37ca68-63d3-47d3-adbb-2c6abe6ab633
| Comm: inproc://192.168.86.116/149707/1 | Workers: 1 |
| Dashboard: http://192.168.86.116:8787/status | Total threads: 14 |
| Started: Just now | Total memory: 38.76 GiB |
Workers
Worker: 0
| Comm: inproc://192.168.86.116/149707/4 | Total threads: 14 |
| Dashboard: http://192.168.86.116:34953/status | Memory: 38.76 GiB |
| Nanny: None | |
| Local directory: /tmp/dask-scratch-space/worker-a59lxz2m | |
Open the dask dashboard at the link above to monitor task progress.
Organize file paths#
Set up some string variables for directory paths. We need to pass xr.open_mfdataset() a list of all files to read in. Currently, the file structure is organized so that each scene has its own sub-directory within asf_rtcs. Within each sub-directory are several files - we want to extract the GeoTIFF files containing RTC imagery for the VV and VH polarizations for each scene as well as the layover-shadow mask for each scene. The function extract_fnames() takes a path to the directory containing the sub-directories for all scenes and returns a list of the filenames for VV-polarization GeoTIFF files, VH-polarization GeoTIFF files and layover-shadow mask GeoTIFF files.
cwd = pathlib.Path.cwd()
tutorial2_dir = pathlib.Path(cwd).parent
timeseries_type = "full"
path_to_rtcs = f"sentinel1/data/raster_data/{timeseries_type}_timeseries/asf_rtcs"
# Path to directory holding downloaded data
s1_asf_data = os.path.join(str(cwd.parents[1]), path_to_rtcs)
scenes_ls = os.listdir(s1_asf_data)
def extract_fnames(data_path: str, scene_name: str, variable: str) -> list:
"""
Extract filenames for a specific variable from a given scene directory.
Parameters
----------
data_path : str
Path to the directory containing scene sub-directories.
scene_name : str
Name of the scene sub-directory.
variable : str
Variable type to extract filenames for. Options are 'vv', 'vh', 'ls_map', or 'readme'.
Returns
-------
list
List of filenames matching the specified variable within the scene directory.
"""
# Make list of files within each scene directory in data directory
scene_files_ls = os.listdir(os.path.join(data_path, scene_name))
if variable in ["vv", "vh"]:
scene_files = [fname for fname in scene_files_ls if fname.endswith(f"_{variable.upper()}.tif")]
elif variable == "ls_map":
scene_files = [fname for fname in scene_files_ls if fname.endswith(f"_ls_map.tif")]
elif variable == "readme":
scene_files = [file for file in scene_files_ls if file.endswith("README.md.txt")]
return scene_files
print(extract_fnames(s1_asf_data, scenes_ls[0], "vv"))
print(extract_fnames(s1_asf_data, scenes_ls[0], "vh"))
print(extract_fnames(s1_asf_data, scenes_ls[0], "ls_map"))
print(extract_fnames(s1_asf_data, scenes_ls[0], "readme"))
['S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_VV.tif']
['S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_VH.tif']
['S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_ls_map.tif']
['S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7.README.md.txt']
def make_filepath_lists(asf_s1_data_path: str, variable: str) -> tuple:
"""
Generate lists of file paths and acquisition dates for a specific variable from Sentinel-1 RTC data.
Parameters
----------
asf_s1_data_path : str
Path to the directory containing scene sub-directories.
variable : str
Variable type to extract filenames for. Options are 'vv', 'vh', 'ls_map', or 'readme'.
Returns
-------
tuple
A tuple containing two lists:
- fpaths (list of str): List of full file paths for the specified variable.
- dates_ls (list of str): List of acquisition dates corresponding to each file path.
"""
scenes_ls = os.listdir(asf_s1_data_path)
fpaths, dates_ls = [], []
for element in range(len(scenes_ls)):
# Extract filenames of each file of interest
files_of_interest = extract_fnames(asf_s1_data_path, scenes_ls[element], variable)
# Make full path with filename for each variable
path = os.path.join(asf_s1_data_path, scenes_ls[element], files_of_interest[0])
# Extract dates to make sure dates are identical across variable lists
date = pathlib.Path(path).stem.split("_")[2]
dates_ls.append(date)
fpaths.append(path)
return (fpaths, dates_ls)
def create_filenames_dict(rtc_path: str, variables_ls: list) -> dict:
"""
Create a dictionary of filenames for specified variables from Sentinel-1 RTC data.
Parameters
----------
rtc_path : str
Path to the directory containing scene sub-directories.
variables_ls : list
List of variable types to extract filenames for. Options are 'vv', 'vh', 'ls_map', or 'readme'.
Returns
-------
dict
A dictionary where keys are variable names and values are lists of file paths for the specified variables.
Raises
------
AssertionError
If the dates are not identical across all lists or if the length of each variable list is not the same.
"""
keys, filepaths, dates = [], [], []
for variable in variables_ls:
keys.append(variable)
filespaths_list, dates_list = make_filepath_lists(rtc_path, variable)
filepaths.append(filespaths_list)
dates.append(dates_list)
# Make dict of variable names (keys) and associated filepaths
filepaths_dict = dict(zip(keys, filepaths))
# Make sure that dates are identical across all lists
assert all(lst == dates[0] for lst in dates) == True
# Make sure length of each variable list is the same
assert len(list(set([len(v) for k, v in filepaths_dict.items()]))) == 1
return filepaths_dict
filepaths_dict = create_filenames_dict(s1_asf_data, ["vv", "vh", "ls_map", "readme"])
filepaths_dict has a key for each file type:
filepaths_dict.keys()
dict_keys(['vv', 'vh', 'ls_map', 'readme'])
Read files using xr.open_mfdataset()#
The xr.open_mfdataset() function reads multiple files (in a directory or from a list) and combines them to return a single xr.Dataset or xr.DataArray. To use the function, specify parameters such as how the files should be combined as well as any preprocessing to execute on the original files. This example will demonstrate a workflow for using open_mfdataset() to read in three stacks of roughly 100 RTC images.
Here is an example of reading a single file with xr.open_datatset():
ds1 = xr.open_dataset(filepaths_dict["vv"][0])
ds1
<xarray.Dataset> Size: 302MB
Dimensions: (band: 1, x: 9851, y: 7656)
Coordinates:
* band (band) int64 8B 1
* x (x) float64 79kB 4.152e+05 4.152e+05 ... 7.106e+05 7.107e+05
* y (y) float64 61kB 3.137e+06 3.137e+06 ... 2.907e+06 2.907e+06
spatial_ref int64 8B ...
Data variables:
band_data (band, y, x) float32 302MB ...We’re missing a lot of important metadata such as the type of data (VV, VH or ls_map) and the time associated with the observation. This is stored in the filename. We will use a preprocess function to tell Xarray how to parse the filename and organize the related metadata for each file in the list passed to xr.open_mfdataset(). The preprocess function will use parse_fname_metadata(), defined below:
def parse_fname_metadata(input_fname: str) -> dict:
"""Function to extract information from filename and separate into expected variables based on a defined schema."""
# Define schema
schema = {
"sensor": (3, r"S1[A-B]"), # schema for sensor
"beam_mode": (2, r"[A-Z]{2}"), # schema for beam mode
"acq_date": (15, r"[0-9]{8}T[0-9]{6}"), # schema for acquisition date
"pol_orbit": (3, r"[A-Z]{3}"), # schema for polarization + orbit type
"terrain_correction_pixel_spacing": (
5,
r"RTC[0-9]{2}",
), # schema for terrain correction pixel spacing
"processing_software": (
1,
r"[A-Z]{1}",
), # schema for processing software (G = Gamma)
"output_info": (6, r"[a-z]{6}"), # schema for output info
"product_id": (4, r"[A-Z0-9]{4}"), # schema for product id
"prod_type": ((2, 6), (r"[A-Z]{2}", r"ls_map")), # schema for polarization type
}
# Remove prefixs
input_fname = input_fname.split("/")[-1]
# Remove file extension if present
input_fname = input_fname.removesuffix(".tif")
# Split filename string into parts
parts = input_fname.split("_")
# l-s map objects have an extra '_' in the filename. Remove/combine parts so that it matches schema
if parts[-1] == "map":
parts = parts[:-1]
parts[-1] = parts[-1] + "_map"
# Check that number of parts matches expected schema
if len(parts) != len(schema):
raise ValueError(f"Input filename does not match schema of expected format: {parts}")
# Create dict to store parsed data
parsed_data = {}
# Iterate through parts and schema
for part, (name, (length_options, pattern_options)) in zip(parts, schema.items()):
# In the schema we defined, items have an int for length or a tuple (when there is more than one possible length)
# Make the int lengths into tuples
if isinstance(length_options, int):
length_options = (length_options,)
# Same as above for patterns
if isinstance(pattern_options, str):
pattern_options = (pattern_options,)
# Check that each length of each part matches expected length from schema
if len(part) not in length_options:
raise ValueError(f"Part {part} does not have expected length {len(part)}")
# Check that each part matches expected pattern from schema
if not any(re.fullmatch(pattern, part) for pattern in pattern_options):
raise ValueError(f"Part {part} does not match expected patterns {pattern_options}")
# Special handling of a part (pol orbit) that has 3 types of metadata
if name == "pol_orbit":
parsed_data.update(
{
"polarization_type": part[:1], # Single (S) or Dual (D) pol
"primary_polarization": part[1:2], # Primary polarization (H or V)
"orbit_type": part[-1], # Precise (p), Restituted (r) or Original predicted (o)
}
)
# Format string acquisition date as a datetime time stamp
elif name == "acq_date":
parsed_data[name] = pd.to_datetime(part, format="%Y%m%dT%H%M%S")
# Expand multiple variables stored in output_info string part
elif name == "output_info":
output_info_keys = [
"output_type",
"output_unit",
"unmasked_or_watermasked",
"notfiltered_or_filtered",
"area_or_clipped",
"deadreckoning_or_demmatch",
]
output_info_values = [part[0], part[1], part[2], part[3], part[4], part[-1]]
parsed_data.update(dict(zip(output_info_keys, output_info_values)))
else:
parsed_data[name] = part
# Because we have already addressed product type in the variable names
prod_type = parsed_data.pop("prod_type")
return parsed_data
Now that we’ve written a function to create a dictionary of relevant metadata extracted from a filename, we need to use it within a preprocess function. Preprocess functions ingest and return xr.Dataset or xr.DataArray objects; in other words, they act on each unit passed to open_mfdataset(). Your preprocess function should contain all of the steps that must be applied to a single file/xr.Dataset object to end up with an analysis-ready cube. The preprocess function below:
Renames the data variable from
'band_data'to a descriptive name,Extracts the file name associated with the
xr.Dataset(from the object’s encoding),Parses information stored in the filename and adds it as an
attrsdictionary,Extracts coordinate reference system metadata,
Reshapes the dataset into a 3-dimensional
x,y,timecube
def preprocess(ds_orig: xr.DataArray, variable: str):
"""function that should return an xarray object with time dimension and associated metadata given a path to a single RTC scene, if its dualpol will have multiple bands, currently just as 2 data arrays but could merge.
goal would be to apply this a list of directories for different RTC products, return cube along time dimension - I think?
- for concatenating, would need to check footprints and only take products with the same footprint, or subset them all to a common AOI?
"""
# fname_ls = []
ds = ds_orig.copy()
ds = ds.rename({"band_data": variable}).squeeze()
# vv_fn = da_orig.encoding['source'][113:]
vh_fn = os.path.basename(ds_orig["band_data"].encoding["source"])
# print('fname: ', vv_fn)
attrs_dict = parse_fname_metadata(vh_fn)
# link the strings for each of the above variables to their full names (from README, commented above)
# eg if output_type=g, should read 'gamma'
ds.attrs = attrs_dict
utm_zone = ds.spatial_ref.attrs["crs_wkt"][17:29]
epsg_code = ds.spatial_ref.attrs["crs_wkt"][589:594]
ds.attrs["utm_zone"] = utm_zone
ds.attrs["epsg_code"] = f"EPSG:{epsg_code}"
date = ds.attrs["acq_date"]
ds = ds.assign_coords({"acq_date": date})
ds = ds.expand_dims("acq_date")
ds = ds.drop_duplicates(dim=["x", "y"])
return ds
Create the wrapper function for each variable:
preprocess_vh_partial = partial(preprocess, variable="vh")
preprocess_vv_partial = partial(preprocess, variable="vv")
preprocess_ls_partial = partial(preprocess, variable="ls_map")
An example of complicated chunking#
First, let’s call xr.open_mfdataset() with the argument chunks='auto'. This will read in a dask array where ideal chunk sizes are selected based off the array size, it will attempt to have chunk sizes where bytes are equal to the configuration value for array chunk size. More about that here. You can check the configuration value for an array chunk size with the code below:
dask.config.get("array.chunk-size")
'128MiB'
asf_vh = xr.open_mfdataset(
paths=filepaths_dict["vh"],
preprocess=preprocess_vh_partial,
chunks="auto",
engine="rasterio",
data_vars="minimal",
coords="minimal",
concat_dim="acq_date",
combine="nested",
parallel=True,
)
asf_vv = xr.open_mfdataset(
paths=filepaths_dict["vv"],
preprocess=preprocess_vv_partial,
chunks="auto",
engine="rasterio",
data_vars="minimal",
coords="minimal",
concat_dim="acq_date",
combine="nested",
parallel=True,
)
asf_ls = xr.open_mfdataset(
paths=filepaths_dict["ls_map"],
preprocess=preprocess_ls_partial,
chunks="auto",
engine="rasterio",
data_vars="minimal",
coords="minimal",
concat_dim="acq_date",
combine="nested",
parallel=True,
)
If we take a look at the asf_vh object we just created (click the stack icon on the right of the vh variable tab), we see that the chunking along the x and y dimensions is quite complicated. This isn’t ideal because it can create problems like excessive communication between workers (and a lot of memory usage) down the line when we perform non-lazy operations. It seems like the 'auto' chunking is applied to the top layer (file) of the dataset, but because the spatial footprint of each file is not the same, the ‘auto’ chunking does not persist through layers and we end up with the funky layout in the object above.
Ensure that each of the three objects have the same coordinate dimensions so that they can be combined:
assert asf_vh.dims == asf_vv.dims == asf_ls.dims
assert asf_ls.acq_date.equals(asf_vv.acq_date)
assert asf_ls.acq_date.equals(asf_vh.acq_date)
assert asf_vh.x.equals(asf_vv.x)
assert asf_vh.x.equals(asf_ls.x)
assert asf_vh.y.equals(asf_vv.y)
assert asf_vh.y.equals(asf_ls.y)
Merge the VH, VV and layover-shadow mask objects into one xr.Dataset:
asf_ds = xr.Dataset({"vv": asf_vv.vv, "vh": asf_vh.vh, "ls": asf_ls.ls_map})
asf_ds
<xarray.Dataset> Size: 289GB
Dimensions: (x: 17452, y: 13379, acq_date: 103)
Coordinates:
* x (x) float64 140kB 3.833e+05 3.833e+05 ... 9.068e+05 9.068e+05
* y (y) float64 107kB 2.907e+06 2.907e+06 ... 3.309e+06 3.309e+06
* acq_date (acq_date) datetime64[ns] 824B 2022-02-14T12:13:53 ... 2022-...
band int64 8B 1
spatial_ref int64 8B 0
Data variables:
vv (acq_date, y, x) float32 96GB dask.array<chunksize=(1, 3753, 4861), meta=np.ndarray>
vh (acq_date, y, x) float32 96GB dask.array<chunksize=(1, 3753, 4861), meta=np.ndarray>
ls (acq_date, y, x) float32 96GB dask.array<chunksize=(1, 3753, 4861), meta=np.ndarray>If you take a look at the acq_date coordinate, you will see that they are not in order. Let’s sort by the acq_date (time) dimension using xarray .sortby():
asf_ds_sorted = asf_ds.sortby(asf_ds.acq_date)
asf_ds_sorted
<xarray.Dataset> Size: 289GB
Dimensions: (x: 17452, y: 13379, acq_date: 103)
Coordinates:
* x (x) float64 140kB 3.833e+05 3.833e+05 ... 9.068e+05 9.068e+05
* y (y) float64 107kB 2.907e+06 2.907e+06 ... 3.309e+06 3.309e+06
* acq_date (acq_date) datetime64[ns] 824B 2021-05-02T12:14:14 ... 2022-...
band int64 8B 1
spatial_ref int64 8B 0
Data variables:
vv (acq_date, y, x) float32 96GB dask.array<chunksize=(1, 3753, 4861), meta=np.ndarray>
vh (acq_date, y, x) float32 96GB dask.array<chunksize=(1, 3753, 4861), meta=np.ndarray>
ls (acq_date, y, x) float32 96GB dask.array<chunksize=(1, 3753, 4861), meta=np.ndarray>How large is this object?
asf_ds_sorted.nbytes / 1e9
288.594268176
Clip stack to AOI using rioxarray.clip()#
This is a pretty unwieldy object. Let’s subset it down to just the area we want to focus on. Read in the following GeoJSON to clip the dataset to the area of interest we’ll be using in this tutorial. This AOI covers a region in the central Himalaya near the Chinese border in the Sikkim region of India. We will use the rioxarray.clip() function which you can read more about here.
pc_aoi = gpd.read_file("https://github.com/e-marshall/sentinel1_rtc/raw/main/hma_rtc_aoi.geojson")
pc_aoi
| geometry | |
|---|---|
| 0 | POLYGON ((619420.000 3089790.000, 628100.000 3... |
pc_aoi.explore()
Clip the object:
asf_clip = asf_ds_sorted.rio.clip(pc_aoi.geometry, pc_aoi.crs)
Check the size of the clipped object:
asf_clip.nbytes / 1e9 # units are GB
0.141948568
asf_clip
<xarray.Dataset> Size: 142MB
Dimensions: (x: 290, y: 396, acq_date: 103)
Coordinates:
* x (x) float64 2kB 6.194e+05 6.195e+05 ... 6.281e+05 6.281e+05
* y (y) float64 3kB 3.09e+06 3.09e+06 ... 3.102e+06 3.102e+06
* acq_date (acq_date) datetime64[ns] 824B 2021-05-02T12:14:14 ... 2022-...
band int64 8B 1
spatial_ref int64 8B 0
Data variables:
vv (acq_date, y, x) float32 47MB dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
vh (acq_date, y, x) float32 47MB dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
ls (acq_date, y, x) float32 47MB dask.array<chunksize=(1, 396, 290), meta=np.ndarray># If we called compute right now it would kill the kernel
# asf_clip_load = asf_clip.compute()
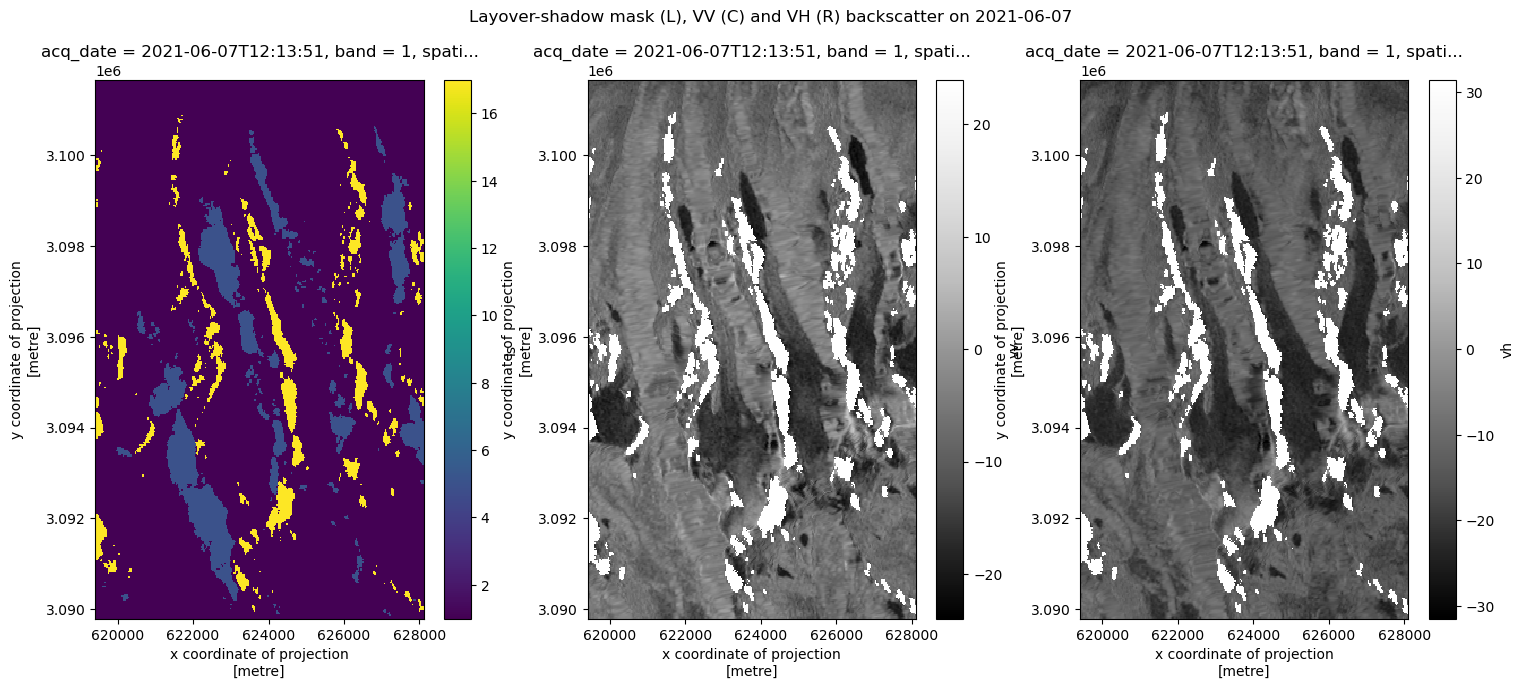
The dataset is published as gamma-nought values on the power scale. This is useful for performing statistical analysis but not always for visualization. We convert to decibel scale (sigma-nought) in order to visualize variability in backscatter more easily. Read more about scales used to represent SAR data here.
def power_to_db(input_arr):
return 10 * np.log10(np.abs(input_arr))
fig, axs = plt.subplots(ncols=3, figsize=(18, 7))
asf_clip.isel(acq_date=10).ls.plot(ax=axs[0])
power_to_db(asf_clip.isel(acq_date=10).vv).plot(ax=axs[1], cmap=plt.cm.Greys_r)
power_to_db(asf_clip.isel(acq_date=10).vh).plot(ax=axs[2], cmap=plt.cm.Greys_r)
fig.suptitle(
f"Layover-shadow mask (L), VV (C) and VH (R) backscatter on {str(asf_clip.isel(acq_date=10).acq_date.data)[:-19]}"
);
Wrap-up#
The xr.open_mfdataset() approach is very useful for reading in large amounts of data from a number of distributed files very efficiently. It’s especially helpful that the preprocess() function allows a large degree of customization for how the data is read in, and that the appropriate metadata is preserved as individual xr.DataArrays are organized into data cubes and we have to do very little further work of organizing metadata.
However, due to the nature of this stack of data, where each time-element of the stack covers a common region of interest but does not have a uniform spatial footprint, this approach is not very computationally efficient.