4.1 Read Sentinel-1 data processed by ASF#
This notebook demonstrates working with Sentinel-1 RTC imagery that has been processed on the ASF On-Demand server and downloaded locally.
The downloaded time series of Sentinel-1 imagery is very large. We demonstrate strategies for reading data of this nature into memory by creating a virtual copy of the data.
A. Prepare to read data into memory
Build lists of file names and paths needed for VRT objects
Create VRT objects
Take a look at chunking
Concepts
Understand local file storage and create virtual datasets from locally stored files
Read larger-than-memory data into memory
Use VRT files to create 3-d Xarray data cubes from stack of 2-d image files
Techniques
Create GDAL VRT files.
TODO add these
Expand the next cell to see specific packages used in this notebook and relevant system and version information.
Show code cell source
# %xmode minimal
import os
import pathlib
import xarray as xr
cwd = pathlib.Path.cwd()
tutorial2_dir = pathlib.Path(cwd).parent
A. Prepare to read data into memory#
1) Build lists of file names and paths needed for VRT objects#
After the data is extracted from the compressed files, we have a directory containing sub-directories for each Sentinel-1 image acquisition (scene). Within each sub-directory are all of the files associated with that scene. For more information about the files contained in each directory, see this section of the ASF Sentinel-1 RTC Product Guide.
The directory should look like the diagram below:
.
└── s1_asf_data
├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_E1E7
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_VH.tif.xml
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_rgb.kmz
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_shape.prj
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7.png.aux.xml
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7.README.md.txt
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_rgb.png.aux.xml
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_rgb.png
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_VV.tif.xml
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7.png
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_rgb.png.xml
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7.png.xml
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_shape.shp
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_VH.tif
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7.log
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_VV.tif
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7.kmz
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_shape.dbf
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_ls_map.tif.xml
│ ├── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_shape.shx
│ └── S1A_IW_20220214T121353_DVP_RTC30_G_gpuned_51E7_ls_map.tif
└── S1A_IW_20210505T000307_DVP_RTC30_G_gpuned_54B1
└── ...
To build GDAL VRT files, we need to pass a list of the input datasets. This means that we need to extract the file path to every file associated with each variable (VV, VH and L-S).
Attention
If you are following along on your own computer, be sure to specify two things:
Set
path_to_rtcsto the location of the downloaded files on your own computer.Set
timeseries_typeto'full'or'subset'depending on if you are using the full time series (103 scenes) or the subset time series (5 scenes).
timeseries_type = "subset"
path_to_rtcs = f"data/raster_data/{timeseries_type}_timeseries/asf_rtcs"
# Path to directory holding downloaded data
s1_asf_data = pathlib.Path(cwd.parent, path_to_rtcs)
Make a list of all scenes:
scenes_ls = os.listdir(s1_asf_data)
scenes_ls
['S1A_IW_20210514T121349_DVP_RTC30_G_gpuned_971C',
'S1A_IW_20210502T121414_DVP_RTC30_G_gpuned_1424',
'S1A_IW_20210509T120542_DVP_RTC30_G_gpuned_8A4F',
'S1A_IW_20210505T000307_DVP_RTC30_G_gpuned_54B1',
'S1A_IW_20210514T121414_DVP_RTC30_G_gpuned_FA4F']
The below function makes a list of all file names across the individual scene directory for VV, VH, ls_map and README files.
def extract_fnames(data_path: str, scene_name: str, variable: str):
# Make list of files within each scene directory in data directory
scene_files_ls = os.listdir(os.path.join(data_path, scene_name))
if variable in ["vv", "vh"]:
scene_files = [fname for fname in scene_files_ls if fname.endswith(f"_{variable.upper()}.tif")]
elif variable == "ls_map":
scene_files = [fname for fname in scene_files_ls if fname.endswith("_ls_map.tif")]
elif variable == "readme":
scene_files = [file for file in scene_files_ls if file.endswith("README.md.txt")]
return scene_files
Below is the output of extract_fnames() for two one scene directory. Note that os.listdir() does not preserve the order of the subdirectories as listed on disk. This is okay because we will ensure that the files are sorted in chronological order later.
print(extract_fnames(s1_asf_data, scenes_ls[0], "vv"))
print(extract_fnames(s1_asf_data, scenes_ls[0], "vh"))
print(extract_fnames(s1_asf_data, scenes_ls[0], "ls_map"))
print(extract_fnames(s1_asf_data, scenes_ls[0], "readme"))
['S1A_IW_20210514T121349_DVP_RTC30_G_gpuned_971C_VV.tif']
['S1A_IW_20210514T121349_DVP_RTC30_G_gpuned_971C_VH.tif']
['S1A_IW_20210514T121349_DVP_RTC30_G_gpuned_971C_ls_map.tif']
['S1A_IW_20210514T121349_DVP_RTC30_G_gpuned_971C.README.md.txt']
We need to attach the filenames to the path of each file so that we end up with a list of the full paths to the VV and VH band imagery, layover-shadow maps and README files. Within this step, we will also add checks to ensure that the process is doing what we want, which is to create lists of filepaths for each character with the same order across lists so that we can combine the lists into a multivariate time series. As noted above, it is okay that the lists are not in chronological order, as long as they are in the same order across variables.
def make_filepath_lists(asf_s1_data_path: str, variable: str):
"""For a single variable (vv, vh, ls_map or readme), make list of
full filepath for each file in time series. Also return dates to ensure
extraction happens in correct order for each variable
Return tuple with form (filepaths list, dates list)"""
scenes_ls = os.listdir(asf_s1_data_path)
fpaths, dates_ls = [], []
for element in range(len(scenes_ls)):
# Extract filenames of each file of interest
files_of_interest = extract_fnames(asf_s1_data_path, scenes_ls[element], variable)
# Make full path with filename for each variable
path = os.path.join(asf_s1_data_path, scenes_ls[element], files_of_interest[0])
# extract dates to make sure dates are identical across variable lists
date = pathlib.Path(path).stem.split("_")[2]
dates_ls.append(date)
fpaths.append(path)
return (fpaths, dates_ls)
def create_filenames_dict(rtc_path, variables_ls):
keys, filepaths, dates = [], [], []
for variable in variables_ls:
keys.append(variable)
filespaths_list, dates_list = make_filepath_lists(rtc_path, variable)
filepaths.append(filespaths_list)
dates.append(dates_list)
# make dict of variable names (keys) and associated filepaths
filepaths_dict = dict(zip(keys, filepaths))
# make sure that dates are identical across all lists
assert all(lst == dates[0] for lst in dates) == True
# make sure length of each variable list is the same
assert len(list(set([len(v) for k, v in filepaths_dict.items()]))) == 1
# make dict of variable names (keys) and associated filepaths
filepaths_dict = dict(zip(keys, filepaths))
return filepaths_dict
filepaths_dict = create_filenames_dict(s1_asf_data, ["vv", "vh", "ls_map", "readme"])
filepaths_dict has a key for each file type:
filepaths_dict.keys()
dict_keys(['vv', 'vh', 'ls_map', 'readme'])
The values are the paths to each file.
2) Create VRT objects#
We will be using the gdalbuildvrt command. You can find out more about it here. This command creates a virtual GDAL dataset given a list of other GDAL datasets (the Sentinel-1 scenes). gdalbuildvrt can make a VRT that either tiles the listed files into a large spatial mosaic, or places them each in a separate band of the VRT. Because we are dealing with a temporal stack of images we want to use the -separate flag to place each file into a band of the VRT.
Here is where we use the list of file paths we created above. For each variable, write the list of file paths to a .txt file which is then passed to gdalbuildvrt.
def create_vrt_object(fpaths_dict: dict, variable: str, timeseries_type: str):
"""Function to create VRT files for each variable given a
list of file paths for that variable."""
full_timeseries_data = "../data/raster_data/full_timeseries"
subset_timeseries_data = "../data/raster_data/subset_timeseries"
if timeseries_type == "full":
data_path = full_timeseries_data
elif timeseries_type == "subset":
data_path = subset_timeseries_data
# switch 'ls' to match variable name in dict
if variable == "ls":
variable = "ls_map"
# Create text files subdir
pathlib.Path(data_path, "txt_files").mkdir(parents=True, exist_ok=True)
# Write file paths to txt file
fpath_input_txt = pathlib.Path(data_path, f"txt_files/s1_{variable}_fpaths.txt")
# Specify location of vrt file- note that we use
# tutorial2_dir path defined at top of notebook
output_vrt_path = pathlib.Path(data_path, f"vrt_files/s1_stack_{variable}.vrt")
# Write file paths to txt file
with open(fpath_input_txt, "w") as fp:
for item in fpaths_dict[f"{variable}"]:
fp.write(f"{item}\n")
!gdalbuildvrt -separate -input_file_list {fpath_input_txt} {output_vrt_path}
create_vrt_object(filepaths_dict, "vv", timeseries_type)
create_vrt_object(filepaths_dict, "vh", timeseries_type)
create_vrt_object(filepaths_dict, "ls", timeseries_type)
0...10...20...30...40...50...60...70...80...90...100 - done.
0...10...20
...30...40...50...60...70...80...90...100 - done.
0...10...20
...30...40...50...60...70...80...90...100 - done.
B. Read data#
timeseries_type
'subset'
ds_vv = xr.open_dataset(
f"../data/raster_data/{timeseries_type}_timeseries/vrt_files/s1_stack_vv.vrt",
chunks="auto",
)
ds_vh = xr.open_dataset(
f"../data/raster_data/{timeseries_type}_timeseries/vrt_files/s1_stack_vh.vrt",
chunks="auto",
)
ds_ls = xr.open_dataset(
f"../data/raster_data/{timeseries_type}_timeseries/vrt_files/s1_stack_ls_map.vrt",
chunks="auto",
)
Building the VRT object assigns every object in the .txt file to a different band. In doing this, we lose the metadata that is associated with the files. The next notebook walks through the steps of reconstructing necessary metadata stored in file names and auxiliary files and attaching it to the Xarray objects read from VRTs.
1) Take a look at chunking#
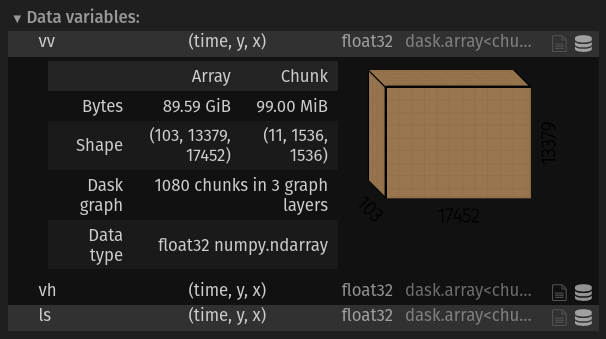
Each variable in ds has a total shape of (103, 13379, 1742) and is 89.59 GB. It is chunked so that each chunk is (11, 1536, 1536) and 99 MB, with 1080 total chunks per variable.
Depending on your use-case, you may want to adjust the chunking of the object. For example, if you are interested in analyzing variability along the temporal dimension, it might make sense to re-chunk the dataset so that operations along the that dimension are more easily parallelized. For more detail, see the chunking discussion in Relevant Concepts and the Parallel Computing with Dask section of the Xarray documentation.
Conclusion#
This notebook demonstrated reading large data into memory by creating a virtual dataset that references that full dataset without directly reading it. However, we also saw that reading the data in this way produces an object that lacks important metadata. The next notebook will go through the steps of locating and adding relevant metadata to the backscatter data cubes read in this notebook.