3.3 Handling raster and vector data#
Introduction#
In the previous notebook, we worked through challenges that can be associated with working with larger-than-memory datasets. Here, we’ll start where we left off, with a dataset that is organized in chronological order, and has a chunking strategy applied.
So far, we’ve been looking at ice velocity data generated for vast spatial footprints. What we’re actually interested in is how ice velocity varies within and across individual glaciers. To look at this more closely, we’ll use glacier outlines the Randolph Glacier Inventory to subset the velocity data. Once we’ve done this, we can start to ask questions like how does velocity vary over time and across different seasons, where on the glacier do velocities tend to be fastest and most seasonally active, and how do these patterns vary across different glaciers? The last two notebooks of this tutorial will start to address these questions.
In this notebook, we introduce vector data that describes spatial features that we’re interested in. This notebook will:
Look at how to parse and inspect important geographic metadata.
Project data to match the coordinate reference system (CRS) of another data object.
Join raster and vector data by clipping the raster data cube by the spatial extent of a vector data frame.
Write this clipped raster object to disk.
A. Read data using strategy identified in previous notebook
B. Incorporate glacier outline (vector) data
Read and reproject vector data
Visualize spatial extents of glacier outlines and ITS_LIVE data cube
Crop vector data to spatial extent of raster data
C. Combine raster and vector data
Use vector data to crop raster data
Write clipped raster data cube to disk
Concepts
Understand how to parse and manage coordinate reference system metadata,
Visualize vector dataframes with static and interactive plots,
Spatial subsetting and clipping with vector and raster data,
Write raster data to disk.
Techniques
Expand the next cell to see specific packages used in this notebook and relevant system and version information.
Show code cell source
import cf_xarray
import folium
import geopandas as gpd
import matplotlib.pyplot as plt
import pathlib
import pyproj
import itslive_tools
A. Read data using strategy identified in previous notebook#
Because we will be reading and writing files to disk, create a variable to hold the path to the root directory for this tutorial; we’ll use this later on.
cwd = pathlib.Path.cwd()
tutorial1_dir = pathlib.Path(cwd).parent
# Find url
url = itslive_tools.find_granule_by_point([95.180191, 30.645973])
# Read data cube without Dask
dc = itslive_tools.read_in_s3(url, chunks=None)
# Sort by mid-date
dc = dc.sortby("mid_date")
# Visually check mid-date in chronological order
dc.mid_date
<xarray.DataArray 'mid_date' (mid_date: 47892)> Size: 383kB
array(['1986-09-11T03:31:15.003252992', '1986-10-05T03:31:06.144750016',
'1986-10-21T03:31:34.493249984', ..., '2024-10-29T04:18:09.241024000',
'2024-10-29T04:18:09.241024000', '2024-10-29T04:18:09.241024000'],
dtype='datetime64[ns]')
Coordinates:
mapping <U1 4B ...
* mid_date (mid_date) datetime64[ns] 383kB 1986-09-11T03:31:15.003252992 ....
Attributes:
description: midpoint of image 1 and image 2 acquisition date and time...
standard_name: image_pair_center_date_with_time_separationLike in the last notebook, we want to chunk the dataset and to do that, we need to assign chunk sizes for each dimension:
chunking_dict = {"mid_date": 20000, "y": 10, "x": 10}
dc = dc.chunk(chunking_dict)
dc.chunks
Frozen({'mid_date': (20000, 20000, 7892), 'y': (10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 3), 'x': (10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 3)})
Looks good 👍
B. Incorporate glacier outline (vector) data#
1) Read and reproject vector data#
As discussed in the vector data section of the ‘Tutorial Data’ page, the examples in this tutorial use glacier outlines from the Randolph Glacier Inventory], version 7 (RGI7). We’ll specifically be looking at the ‘South Asia East’ region.
se_asia = gpd.read_parquet("../data/vector_data/rgi7_region15_south_asia_east.parquet")
It is vital to check the CRS, or Coordinate Reference Systems, when combining geospatial data from different sources.
The RGI data are in the EPSG:4326 CRS.
se_asia.crs
<Geographic 2D CRS: EPSG:4326>
Name: WGS 84
Axis Info [ellipsoidal]:
- Lat[north]: Geodetic latitude (degree)
- Lon[east]: Geodetic longitude (degree)
Area of Use:
- name: World.
- bounds: (-180.0, -90.0, 180.0, 90.0)
Datum: World Geodetic System 1984 ensemble
- Ellipsoid: WGS 84
- Prime Meridian: Greenwich
The CRS information for the ITS_LIVE dataset is stored in the mapping array. An easy way to discover this is to use the cf_xarray package and search for the grid_mapping variable if present.
dc.cf
Coordinates:
CF Axes: * X: ['x']
* Y: ['y']
Z, T: n/a
CF Coordinates: longitude, latitude, vertical, time: n/a
Cell Measures: area, volume: n/a
Standard Names: * image_pair_center_date_with_time_separation: ['mid_date']
* projection_x_coordinate: ['x']
* projection_y_coordinate: ['y']
Bounds: n/a
Grid Mappings: universal_transverse_mercator: ['mapping']
Data Variables:
Cell Measures: area, volume: n/a
Standard Names: M11_dr_to_vr_factor: ['M11_dr_to_vr_factor']
M12_dr_to_vr_factor: ['M12_dr_to_vr_factor']
autoRIFT_software_version: ['autoRIFT_software_version']
chip_size_height: ['chip_size_height']
chip_size_width: ['chip_size_width']
conversion_matrix_element_11: ['M11']
conversion_matrix_element_12: ['M12']
floating ice mask: ['floatingice']
granule_url: ['granule_url']
image1_acquition_date: ['acquisition_date_img1']
image1_mission: ['mission_img1']
image1_satellite: ['satellite_img1']
image1_sensor: ['sensor_img1']
image2_acquition_date: ['acquisition_date_img2']
image2_mission: ['mission_img2']
image2_satellite: ['satellite_img2']
image2_sensor: ['sensor_img2']
image_pair_center_date: ['date_center']
image_pair_time_separation: ['date_dt']
interpolated_value_mask: ['interp_mask']
land ice mask: ['landice']
land_ice_surface_velocity: ['v']
land_ice_surface_x_velocity: ['vx']
land_ice_surface_y_velocity: ['vy']
region_of_interest_valid_pixel_percentage: ['roi_valid_percentage']
stable_count_slow: ['stable_count_slow']
stable_count_stationary: ['stable_count_stationary']
stable_shift_flag: ['stable_shift_flag']
va_error: ['va_error']
va_error_modeled: ['va_error_modeled']
va_error_slow: ['va_error_slow']
va_error_stationary: ['va_error_stationary']
va_stable_shift: ['va_stable_shift']
va_stable_shift_slow: ['va_stable_shift_slow']
va_stable_shift_stationary: ['va_stable_shift_stationary']
velocity_error: ['v_error']
vr_error: ['vr_error']
vr_error_modeled: ['vr_error_modeled']
vr_error_slow: ['vr_error_slow']
vr_error_stationary: ['vr_error_stationary']
vr_stable_shift: ['vr_stable_shift']
vr_stable_shift_slow: ['vr_stable_shift_slow']
vr_stable_shift_stationary: ['vr_stable_shift_stationary']
vx_error: ['vx_error']
vx_error_modeled: ['vx_error_modeled']
vx_error_slow: ['vx_error_slow']
vx_error_stationary: ['vx_error_stationary']
vx_stable_shift: ['vx_stable_shift']
vx_stable_shift_slow: ['vx_stable_shift_slow']
vx_stable_shift_stationary: ['vx_stable_shift_stationary']
vy_error: ['vy_error']
vy_error_modeled: ['vy_error_modeled']
vy_error_slow: ['vy_error_slow']
vy_error_stationary: ['vy_error_stationary']
vy_stable_shift: ['vy_stable_shift']
vy_stable_shift_slow: ['vy_stable_shift_slow']
vy_stable_shift_stationary: ['vy_stable_shift_stationary']
Bounds: n/a
Grid Mappings: n/a
cube_crs = pyproj.CRS.from_cf(dc.mapping.attrs)
cube_crs
<Projected CRS: EPSG:32646>
Name: WGS 84 / UTM zone 46N
Axis Info [cartesian]:
- [east]: Easting (metre)
- [north]: Northing (metre)
Area of Use:
- undefined
Coordinate Operation:
- name: UTM zone 46N
- method: Transverse Mercator
Datum: World Geodetic System 1984
- Ellipsoid: WGS 84
- Prime Meridian: Greenwich
This indicates that the data is projected to UTM zone 46N (EPSG:32646).
We choose to reproject the glacier outline to the CRS of the datacube:
# Project rgi data to match itslive
se_asia_prj = se_asia.to_crs(cube_crs)
se_asia_prj.head(3)
| rgi_id | o1region | o2region | glims_id | anlys_id | subm_id | src_date | cenlon | cenlat | utm_zone | ... | zmin_m | zmax_m | zmed_m | zmean_m | slope_deg | aspect_deg | aspect_sec | dem_source | lmax_m | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | RGI2000-v7.0-G-15-00001 | 15 | 15-01 | G078088E31398N | 866850 | 752 | 2002-07-10T00:00:00 | 78.087891 | 31.398046 | 44 | ... | 4662.2950 | 4699.2095 | 4669.4720 | 4671.4253 | 13.427070 | 122.267290 | 4 | COPDEM30 | 173 | POLYGON Z ((-924868.476 3571663.111 0.000, -92... |
| 1 | RGI2000-v7.0-G-15-00002 | 15 | 15-01 | G078125E31399N | 867227 | 752 | 2002-07-10T00:00:00 | 78.123699 | 31.397796 | 44 | ... | 4453.3584 | 4705.9920 | 4570.9473 | 4571.2770 | 22.822983 | 269.669144 | 7 | COPDEM30 | 1113 | POLYGON Z ((-921270.161 3571706.471 0.000, -92... |
| 2 | RGI2000-v7.0-G-15-00003 | 15 | 15-01 | G078128E31390N | 867273 | 752 | 2000-08-05T00:00:00 | 78.128510 | 31.390287 | 44 | ... | 4791.7593 | 4858.6807 | 4832.1836 | 4827.6700 | 15.626262 | 212.719681 | 6 | COPDEM30 | 327 | POLYGON Z ((-921061.745 3570342.665 0.000, -92... |
3 rows × 29 columns
How many glaciers are represented in the dataset?
len(se_asia_prj)
18587
2) Visualize spatial extents of glacier outlines and ITS_LIVE data cube#
In Accessing S3 Data, we defined a function to create a vector object describing the footprint of a raster object; we’ll use that again here.
# Get vector bbox of itslive
bbox_dc = itslive_tools.get_bounds_polygon(dc)
bbox_dc["geometry"]
# Check that all objects have correct crs
assert dc.attrs["projection"] == bbox_dc.crs == se_asia_prj.crs
# Plot the outline of the itslive granule and the rgi dataframe together
fig, ax = plt.subplots()
bbox_dc.plot(ax=ax, facecolor="None", color="red")
se_asia_prj.plot(ax=ax, facecolor="None")
<Axes: >
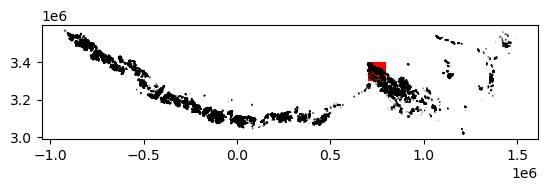
3) Crop vector data to spatial extent of raster data#
The above plot shows the coverage of the vector dataset (se_asia_prj) in black, relative to the extent of the raster dataset (bbox_dc) in red. We use the geopandas .clip() method to subset the RGI polygons to the footprint of the ITS_LIVE datacube.
# Subset rgi to bounds
se_asia_subset = gpd.clip(se_asia_prj, bbox_dc)
se_asia_subset.head()
| rgi_id | o1region | o2region | glims_id | anlys_id | subm_id | src_date | cenlon | cenlat | utm_zone | ... | zmin_m | zmax_m | zmed_m | zmean_m | slope_deg | aspect_deg | aspect_sec | dem_source | lmax_m | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16373 | RGI2000-v7.0-G-15-16374 | 15 | 15-03 | G095930E29817N | 930178 | 752 | 2005-09-08T00:00:00 | 95.929916 | 29.817003 | 46 | ... | 4985.7314 | 5274.0435 | 5142.7660 | 5148.8170 | 27.024134 | 139.048110 | 4 | COPDEM30 | 756 | POLYGON Z ((783110.719 3302487.481 0.000, 7831... |
| 16374 | RGI2000-v7.0-G-15-16375 | 15 | 15-03 | G095925E29818N | 930160 | 752 | 2005-09-08T00:00:00 | 95.925181 | 29.818399 | 46 | ... | 4856.2790 | 5054.9253 | 4929.5560 | 4933.6890 | 44.126980 | 211.518448 | 6 | COPDEM30 | 366 | POLYGON Z ((782511.360 3302381.154 0.000, 7825... |
| 16376 | RGI2000-v7.0-G-15-16377 | 15 | 15-03 | G095915E29820N | 930107 | 752 | 2005-09-08T00:00:00 | 95.914583 | 29.819510 | 46 | ... | 5072.8910 | 5150.6196 | 5108.5020 | 5111.7217 | 23.980000 | 219.341537 | 6 | COPDEM30 | 170 | POLYGON Z ((781619.822 3302305.074 0.000, 7816... |
| 16371 | RGI2000-v7.0-G-15-16372 | 15 | 15-03 | G095936E29819N | 930215 | 752 | 2005-09-08T00:00:00 | 95.935554 | 29.819123 | 46 | ... | 4838.7646 | 5194.8840 | 5001.5117 | 4992.3706 | 25.684517 | 128.737870 | 4 | COPDEM30 | 931 | POLYGON Z ((783420.055 3302493.804 0.000, 7834... |
| 15879 | RGI2000-v7.0-G-15-15880 | 15 | 15-03 | G095459E29807N | 928789 | 752 | 1999-07-29T00:00:00 | 95.459374 | 29.807181 | 46 | ... | 3802.1846 | 4155.1255 | 4000.2695 | 4000.4404 | 28.155806 | 116.148640 | 4 | COPDEM30 | 776 | POLYGON Z ((737667.211 3300277.169 0.000, 7377... |
5 rows × 29 columns
We can use the geopandas .explore() method to interactively look at the RGI7 outlines contained within the ITS_LIVE granule:
m = folium.Map(max_lat=31, max_lon=95, min_lat=29, min_lon=97, location=[30.2, 95.5], zoom_start=8)
bbox_dc.explore(
m=m,
style_kwds={"fillColor": "None", "color": "red"},
legend_kwds={"labels": ["ITS_LIVE granule footprint"]},
)
se_asia_subset.explore(m=m)
folium.LayerControl().add_to(m)
m
/home/emmamarshall/miniforge3/envs/geospatial_datacube_book_env/lib/python3.11/site-packages/folium/features.py:1102: UserWarning: GeoJsonTooltip is not configured to render for GeoJson GeometryCollection geometries. Please consider reworking these features: [{'rgi_id': 'RGI2000-v7.0-G-15-16433', 'o1region': '15', 'o2region': '15-03', 'glims_id': 'G095721E29941N', 'anlys_id': 929520, 'subm_id': 752, 'src_date': '2005-09-08T00:00:00', 'cenlon': 95.7211016152286, 'cenlat': 29.940902187781784, 'utm_zone': 46, 'area_km2': 0.340954350813452, 'primeclass': 0, 'conn_lvl': 0, 'surge_type': 0, 'term_type': 9, 'glac_name': None, 'is_rgi6': 0, 'termlon': 95.72222864596793, 'termlat': 29.937137080413784, 'zmin_m': 4657.792, 'zmax_m': 5049.5625, 'zmed_m': 4825.1104, 'zmean_m': 4839.4185, 'slope_deg': 23.704372, 'aspect_deg': 145.20973, 'aspect_sec': 4, 'dem_source': 'COPDEM30', 'lmax_m': 891}, {'rgi_id': 'RGI2000-v7.0-G-15-12194', 'o1region': '15', 'o2region': '15-03', 'glims_id': 'G095869E30315N', 'anlys_id': 929951, 'subm_id': 752, 'src_date': '2005-09-08T00:00:00', 'cenlon': 95.86889789565677, 'cenlat': 30.3147685, 'utm_zone': 46, 'area_km2': 8.797406997273084, 'primeclass': 0, 'conn_lvl': 0, 'surge_type': 0, 'term_type': 9, 'glac_name': None, 'is_rgi6': 0, 'termlon': 95.89518363763428, 'termlat': 30.307036248571297, 'zmin_m': 4642.1445, 'zmax_m': 5278.752, 'zmed_m': 5011.06, 'zmean_m': 4993.9243, 'slope_deg': 12.372513, 'aspect_deg': 81.418945, 'aspect_sec': 3, 'dem_source': 'COPDEM30', 'lmax_m': 4994}, {'rgi_id': 'RGI2000-v7.0-G-15-11941', 'o1region': '15', 'o2region': '15-03', 'glims_id': 'G095301E30377N', 'anlys_id': 928228, 'subm_id': 752, 'src_date': '2007-08-20T00:00:00', 'cenlon': 95.30071978915663, 'cenlat': 30.3770025, 'utm_zone': 46, 'area_km2': 0.267701958906151, 'primeclass': 0, 'conn_lvl': 0, 'surge_type': 0, 'term_type': 9, 'glac_name': None, 'is_rgi6': 0, 'termlon': 95.30345982475616, 'termlat': 30.380097687364806, 'zmin_m': 5475.784, 'zmax_m': 5977.979, 'zmed_m': 5750.727, 'zmean_m': 5759.621, 'slope_deg': 41.069595, 'aspect_deg': 350.3331518173218, 'aspect_sec': 1, 'dem_source': 'COPDEM30', 'lmax_m': 807}] to MultiPolygon for full functionality.
https://tools.ietf.org/html/rfc7946#page-9
warnings.warn(
We can use the above interactive map to select a glacier to look at in more detail below.
However, notice that while the above code correctly produces a plot, it also throws a warning. We’re going to ignore the warning for now, but if you’re interested in a detailed example of how to trouble shoot and resolve this type of warning, check out the appendix
We write bbox_dc to file so that we can use it in the appendix
bbox_dc.to_file("../data/vector_data/bbox_dc.geojson")
C. Combine raster and vector data#
1) Use vector data to crop raster data#
If we want to dig in and analyze this velocity dataset at smaller spatial scales, we first need to subset it. The following section and the next notebook (Exploratory data analysis of a single glacier) will focus on the spatial scale of a single glacier.
The glacier we’ll focus on is in the southeastern region of Tibetan Plateau in the Hengduan mountain range. It was selected in part because it is large enough that we should be able to observe velocity variability within the context of the spatial resolution of the ITS_LIVE dataset but is not so large that it is an extreme outlier for glaciers in the region.
# Select a glacier to subset to
single_glacier_vec = se_asia_subset.loc[se_asia_subset["rgi_id"] == "RGI2000-v7.0-G-15-16257"]
single_glacier_vec
| rgi_id | o1region | o2region | glims_id | anlys_id | subm_id | src_date | cenlon | cenlat | utm_zone | ... | zmin_m | zmax_m | zmed_m | zmean_m | slope_deg | aspect_deg | aspect_sec | dem_source | lmax_m | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16256 | RGI2000-v7.0-G-15-16257 | 15 | 15-03 | G095962E29920N | 930314 | 752 | 2005-09-08T00:00:00 | 95.961972 | 29.920094 | 46 | ... | 4320.7065 | 5937.84 | 5179.605 | 5131.8877 | 22.803406 | 100.811325 | 3 | COPDEM30 | 5958 | POLYGON Z ((788176.951 3315860.842 0.000, 7882... |
1 rows × 29 columns
# Write it to file to that it can be used later
single_glacier_vec.to_file("../data/single_glacier_vec.json", driver="GeoJSON")
Check to see if the ITS_LIVE raster dataset has an assigned CRS attribute. We already know that the data is projected in the correct coordinate reference system (CRS), but the object may not be ‘CRS-aware’ yet (ie. have an attribute specifying its CRS). This is necessary for spatial operations such as clipping and reprojection. If dc doesn’t have a CRS attribute, use rio.write_crs() to assign it. For more detail, see Rioxarray’s CRS Management documentation.
dc.rio.crs
CRS.from_epsg(32646)
Now, use the subset vector data object and Rioxarray’s .clip() method to crop the data cube.
%%time
single_glacier_raster = dc.rio.clip(single_glacier_vec.geometry, single_glacier_vec.crs)
CPU times: user 2.03 s, sys: 72.4 ms, total: 2.1 s
Wall time: 2.1 s
2) Write clipped raster data cube to disk#
We want to use single_glacier_raster in the following notebook without going through all of the steps of creating it again. So, we write the object to file as a Zarr data cube so that we can easily read it into memory when we need it next. However, we’ll see that there are a few steps we must go through before we can successfully write this object.
We first re-chunk the single_glacier_raster into more optimal chunk sizes:
single_glacier_raster = single_glacier_raster.chunk({"mid_date": 20000, "x": 10, "y": 10})
single_glacier_raster
<xarray.Dataset> Size: 3GB
Dimensions: (mid_date: 47892, x: 40, y: 37)
Coordinates:
* mid_date (mid_date) datetime64[ns] 383kB 1986-09-11T03...
* x (x) float64 320B 7.843e+05 ... 7.889e+05
* y (y) float64 296B 3.316e+06 ... 3.311e+06
spatial_ref int64 8B 0
mapping int64 8B 0
Data variables: (12/59)
M11 (mid_date, y, x) float32 284MB dask.array<chunksize=(20000, 10, 10), meta=np.ndarray>
M11_dr_to_vr_factor (mid_date) float32 192kB dask.array<chunksize=(20000,), meta=np.ndarray>
M12 (mid_date, y, x) float32 284MB dask.array<chunksize=(20000, 10, 10), meta=np.ndarray>
M12_dr_to_vr_factor (mid_date) float32 192kB dask.array<chunksize=(20000,), meta=np.ndarray>
acquisition_date_img1 (mid_date) datetime64[ns] 383kB dask.array<chunksize=(20000,), meta=np.ndarray>
acquisition_date_img2 (mid_date) datetime64[ns] 383kB dask.array<chunksize=(20000,), meta=np.ndarray>
... ...
vy_error_modeled (mid_date) float32 192kB dask.array<chunksize=(20000,), meta=np.ndarray>
vy_error_slow (mid_date) float32 192kB dask.array<chunksize=(20000,), meta=np.ndarray>
vy_error_stationary (mid_date) float32 192kB dask.array<chunksize=(20000,), meta=np.ndarray>
vy_stable_shift (mid_date) float32 192kB dask.array<chunksize=(20000,), meta=np.ndarray>
vy_stable_shift_slow (mid_date) float32 192kB dask.array<chunksize=(20000,), meta=np.ndarray>
vy_stable_shift_stationary (mid_date) float32 192kB dask.array<chunksize=(20000,), meta=np.ndarray>
Attributes: (12/19)
Conventions: CF-1.8
GDAL_AREA_OR_POINT: Area
author: ITS_LIVE, a NASA MEaSUREs project (its-live.j...
autoRIFT_parameter_file: http://its-live-data.s3.amazonaws.com/autorif...
datacube_software_version: 1.0
date_created: 25-Sep-2023 22:00:23
... ...
s3: s3://its-live-data/datacubes/v2/N30E090/ITS_L...
skipped_granules: s3://its-live-data/datacubes/v2/N30E090/ITS_L...
time_standard_img1: UTC
time_standard_img2: UTC
title: ITS_LIVE datacube of image pair velocities
url: https://its-live-data.s3.amazonaws.com/datacu...If we try to write single_glacier_raster, eg:
single_glacier_raster.to_zarr('data/itslive/glacier_itslive.zarr', mode='w')
We’ll received an error related to encoding.
The root cause is that the encoding recorded was appropriate for the source dataset, but is not valid anymore given all the transformations we have run up to this point. The easy solution here is to simply call drop_encoding. This will delete any existing encoding isntructions, and have Xarray automatically choose an encoding that will work well for the data. Optimizing the encoding of on-disk data is an advanced topic that we will not cover.
Now, let’s try to write the object as a Zarr group.
single_glacier_raster.drop_encoding().to_zarr("../data/raster_data/single_glacier_itslive.zarr", mode="w")
<xarray.backends.zarr.ZarrStore at 0x7e73b2be8540>
Conclusion#
In this notebook, we read a large object into memory and clipped it to the footprint of a single area of using a large vector dataframe, all the while managing coordinate reference system metadata of both objects. We then saved the clipped raster object to disk so that it can be easily reused. The next notebook demonstrates exploratory data analysis steps using the object we just wrote to disk.