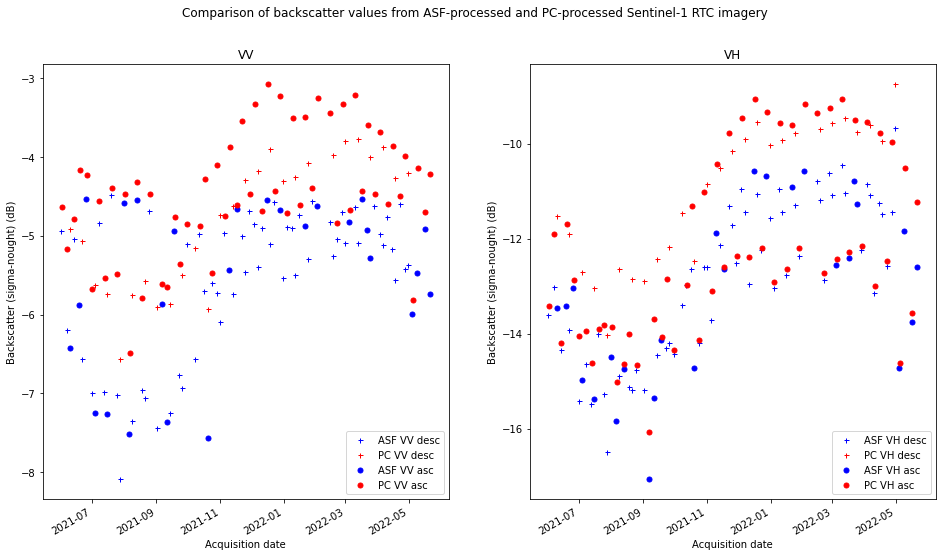
RTC dataset comparison#
Now that we have constructed and organized data cubes for both the ASF and Planetary Computer datasets, it would be useful and informative to compare the two. This notebook will demonstrate steps to ensure a direct comparison between the two datasets as well as use of xarray plotting tools for a preliminary visual comparison.
Learning goals#
Xarray and python techniques:
Conditional selection based on non-dimensional coordinates using
xr.Dataset.where()Subsetting datasets based on dimensional coordinates using
xr.DataArray.isin()Adding dimensional and non-dimensional coordinates to
xr.DatasetobjectsXarray plotting methods
Projecting xarray objects to different grids using
xr.interp_like()
High-level science goals
Comparing and evaluating multiple datasets
Setting up multiple datasets for direct comparisons
Handling differences in spatial resolution
Other useful resources#
These are resources that contain additional examples and discussion of the content in this notebook and more.
How do I… this is very helpful!
Xarray High-level computational patterns discussion of concepts and associated code examples
Parallel computing with dask Xarray tutorial demonstrating wrapping of dask arrays
%xmode minimal
import xarray as xr
import os
import xarray as xr
import rioxarray as rio
import geopandas as gpd
from datetime import datetime
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import dask
%matplotlib inline
Overview#
While the datasets we will be comparing in this notebook are quite similar, they contain important differences related to how they were processed. The product description pages for both datasets contain important information for understanding how they are generated.
Source data#
One important difference is the source data used to generate the RTC images. ASF uses the Single Look Complex (SLC) images that contains both amplitude and phase information for each pixel, is in radar coordinates and has not yet been multi-looked. The Microsoft Planetary Computer RTC imagery is generate from ground range detected images (GRD). GRD data has been detected, multi-looked and projected to ground range.
DEM#
A digital elevation model (DEM) is an important input parameter for RTC processing. ASF RTC processing uses the Copernicus DEM with 30 m resolution. Planetary Computer uses higher resolution DEMs. One effect of the use of different DEMs during RTC processing is the different spatial resolutions of the RTC products: Planetary Computer RTC imagery has a spatial resolution of 10 meters while ASF RTC imagery has a spatial resolution of 30 meters.
Software and setup#
import xarray as xr
import s1_tools
from s1_tools import points2coords
import os
import xarray as xr
import rioxarray as rio
import geopandas as gpd
from datetime import datetime
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import dask
import holoviews as hv
from holoviews import opts
Utility functions#
def power_to_db(input_arr):
return (10*np.log10(np.abs(input_arr)))
def asf_pc_sidebyside(asf_input, pc_input, timestep):
fig, axs = plt.subplots(ncols=2, figsize=(15,10))
power_to_db(asf_input.vv.isel(acq_date=timestep)).plot(ax=axs[0], cmap=plt.cm.Greys_r, label = 'ASF');
power_to_db(pc_input.sel(band='vv').isel(time=timestep)).plot(ax=axs[1], cmap=plt.cm.Greys_r, label = 'PC');
def single_time_mean_compare(asf_input, pc_input, time):
fig, ax = plt.subplots(figsize=(8,8))
power_to_db(asf_input['vv_'].isel(acq_date=time).mean(dim=['x','y'])).plot(ax=ax)
power_to_db(pc_input.sel(band='vv').isel(time=time).mean(dim=['x','y'])).plot(ax=ax, color='red');
Read in prepared data#
We can use the storemagic command %store to retrieve the variable we constructed and saved in a previous notebook, rather than having to create it again. Read more about this here
This let’s us call the ASF dataset (vrt_new) and PC dataset (da_pc)
%store -r vrt_new da_pc
Read in the ASF dataset, which we named vrt_full in the earlier notebook:
vrt_new
<xarray.Dataset>
Dimensions: (acq_date: 95, y: 396, x: 290)
Coordinates: (12/20)
* acq_date (acq_date) datetime64[ns] 2021-05-02 .....
granule_id (acq_date) <U67 'S1A_IW_SLC__1SDV_20210...
* x (x) float64 6.194e+05 ... 6.281e+05
* y (y) float64 3.102e+06 ... 3.09e+06
spatial_ref int64 0
sensor (acq_date) <U3 'S1A' 'S1A' ... 'S1A' 'S1A'
... ...
filtered (acq_date) <U1 'n' 'n' 'n' ... 'n' 'n' 'n'
area (acq_date) <U1 'e' 'e' 'e' ... 'e' 'e' 'e'
product_id (acq_date) <U4 '748F' '0D1E' ... 'BD36'
acq_hour (acq_date) int64 0 12 12 12 12 ... 0 0 0 0
orbital_dir (acq_date) <U4 'asc' 'desc' ... 'asc'
data_take_id (acq_date) <U6 '047321' ... '052C00'
Data variables:
vv (acq_date, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
vh (acq_date, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
ls (acq_date, y, x) float64 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>- acq_date: 95
- y: 396
- x: 290
- acq_date(acq_date)datetime64[ns]2021-05-02 ... 2022-05-21
array(['2021-05-02T00:00:00.000000000', '2021-05-05T00:00:00.000000000', '2021-05-09T00:00:00.000000000', '2021-05-14T00:00:00.000000000', '2021-05-17T00:00:00.000000000', '2021-05-21T00:00:00.000000000', '2021-05-26T00:00:00.000000000', '2021-05-29T00:00:00.000000000', '2021-06-02T00:00:00.000000000', '2021-06-07T00:00:00.000000000', '2021-06-10T00:00:00.000000000', '2021-06-14T00:00:00.000000000', '2021-06-19T00:00:00.000000000', '2021-06-22T00:00:00.000000000', '2021-06-26T00:00:00.000000000', '2021-07-01T00:00:00.000000000', '2021-07-04T00:00:00.000000000', '2021-07-08T00:00:00.000000000', '2021-07-16T00:00:00.000000000', '2021-07-20T00:00:00.000000000', '2021-07-25T00:00:00.000000000', '2021-07-28T00:00:00.000000000', '2021-08-01T00:00:00.000000000', '2021-08-06T00:00:00.000000000', '2021-08-09T00:00:00.000000000', '2021-08-13T00:00:00.000000000', '2021-08-18T00:00:00.000000000', '2021-08-21T00:00:00.000000000', '2021-08-25T00:00:00.000000000', '2021-08-30T00:00:00.000000000', '2021-09-02T00:00:00.000000000', '2021-09-06T00:00:00.000000000', '2021-09-11T00:00:00.000000000', '2021-09-14T00:00:00.000000000', '2021-09-18T00:00:00.000000000', '2021-09-23T00:00:00.000000000', '2021-09-26T00:00:00.000000000', '2021-09-30T00:00:00.000000000', '2021-10-05T00:00:00.000000000', '2021-10-08T00:00:00.000000000', '2021-10-12T00:00:00.000000000', '2021-10-17T00:00:00.000000000', '2021-10-20T00:00:00.000000000', '2021-10-24T00:00:00.000000000', '2021-10-29T00:00:00.000000000', '2021-11-01T00:00:00.000000000', '2021-11-05T00:00:00.000000000', '2021-11-10T00:00:00.000000000', '2021-11-13T00:00:00.000000000', '2021-11-17T00:00:00.000000000', '2021-11-22T00:00:00.000000000', '2021-11-25T00:00:00.000000000', '2021-11-29T00:00:00.000000000', '2021-12-04T00:00:00.000000000', '2021-12-07T00:00:00.000000000', '2021-12-11T00:00:00.000000000', '2021-12-16T00:00:00.000000000', '2021-12-19T00:00:00.000000000', '2021-12-23T00:00:00.000000000', '2021-12-28T00:00:00.000000000', '2021-12-31T00:00:00.000000000', '2022-01-04T00:00:00.000000000', '2022-01-09T00:00:00.000000000', '2022-01-12T00:00:00.000000000', '2022-01-16T00:00:00.000000000', '2022-01-21T00:00:00.000000000', '2022-01-24T00:00:00.000000000', '2022-01-28T00:00:00.000000000', '2022-02-02T00:00:00.000000000', '2022-02-05T00:00:00.000000000', '2022-02-09T00:00:00.000000000', '2022-02-14T00:00:00.000000000', '2022-02-17T00:00:00.000000000', '2022-02-21T00:00:00.000000000', '2022-02-26T00:00:00.000000000', '2022-03-01T00:00:00.000000000', '2022-03-05T00:00:00.000000000', '2022-03-10T00:00:00.000000000', '2022-03-13T00:00:00.000000000', '2022-03-17T00:00:00.000000000', '2022-03-22T00:00:00.000000000', '2022-03-25T00:00:00.000000000', '2022-03-29T00:00:00.000000000', '2022-04-03T00:00:00.000000000', '2022-04-06T00:00:00.000000000', '2022-04-10T00:00:00.000000000', '2022-04-15T00:00:00.000000000', '2022-04-18T00:00:00.000000000', '2022-04-22T00:00:00.000000000', '2022-04-27T00:00:00.000000000', '2022-04-30T00:00:00.000000000', '2022-05-04T00:00:00.000000000', '2022-05-09T00:00:00.000000000', '2022-05-16T00:00:00.000000000', '2022-05-21T00:00:00.000000000'], dtype='datetime64[ns]') - granule_id(acq_date)<U67'S1A_IW_SLC__1SDV_20210502T12141...
- description :
- source granule ID for ASF-processed S1 RTC imagery, extracted from README files for each scene
array(['S1A_IW_SLC__1SDV_20210502T121414_20210502T121441_037709_047321_900F', 'S1A_IW_SLC__1SDV_20210505T000307_20210505T000334_037745_047463_4DEF', 'S1A_IW_SLC__1SDV_20210509T120542_20210509T120609_037811_047676_9EBD', 'S1A_IW_SLC__1SDV_20210514T121349_20210514T121416_037884_047898_4702', 'S1A_IW_SLC__1SDV_20210517T000308_20210517T000335_037920_0479A9_2A18', 'S1A_IW_SLC__1SDV_20210521T120543_20210521T120609_037986_047BBD_1CA0', 'S1A_IW_SLC__1SDV_20210526T121350_20210526T121417_038059_047DE5_0E67', 'S1A_IW_SLC__1SDV_20210529T000309_20210529T000336_038095_047EF4_5449', 'S1A_IW_SLC__1SDV_20210602T120543_20210602T120610_038161_0480FD_893F', 'S1A_IW_SLC__1SDV_20210607T121351_20210607T121418_038234_048318_9B64', 'S1A_IW_SLC__1SDV_20210610T000310_20210610T000337_038270_04841E_601A', 'S1A_IW_SLC__1SDV_20210614T120544_20210614T120611_038336_04862F_337B', 'S1A_IW_SLC__1SDV_20210619T121352_20210619T121419_038409_04884C_1DBA', 'S1A_IW_SLC__1SDV_20210622T000310_20210622T000337_038445_04895A_842E', 'S1A_IW_SLC__1SDV_20210626T120545_20210626T120612_038511_048B6C_9585', 'S1A_IW_SLC__1SDV_20210701T121352_20210701T121419_038584_048D87_F95C', 'S1A_IW_SLC__1SDV_20210704T000311_20210704T000338_038620_048E99_1BAA', 'S1A_IW_SLC__1SDV_20210708T120545_20210708T120612_038686_0490AD_DD9E', 'S1A_IW_SLC__1SDV_20210716T000312_20210716T000339_038795_0493DB_53CD', 'S1A_IW_SLC__1SDV_20210720T120546_20210720T120613_038861_0495EC_C9D4', ... 'S1A_IW_SLC__1SDV_20220305T120545_20220305T120612_042186_0506F7_1601', 'S1A_IW_SLC__1SDV_20220310T121353_20220310T121420_042259_050962_068B', 'S1A_IW_SLC__1SDV_20220313T000311_20220313T000338_042295_050AA2_2909', 'S1A_IW_SLC__1SDV_20220317T120545_20220317T120612_042361_050CE5_4CCF', 'S1A_IW_SLC__1SDV_20220322T121353_20220322T121420_042434_050F59_A1B6', 'S1A_IW_SLC__1SDV_20220325T000312_20220325T000339_042470_051096_BA6F', 'S1A_IW_SLC__1SDV_20220329T120546_20220329T120613_042536_0512D7_425A', 'S1A_IW_SLC__1SDV_20220403T121353_20220403T121420_042609_05154A_3FE7', 'S1A_IW_SLC__1SDV_20220406T000312_20220406T000339_042645_051682_4B40', 'S1A_IW_SLC__1SDV_20220410T120546_20220410T120613_042711_0518C0_3349', 'S1A_IW_SLC__1SDV_20220415T121353_20220415T121420_042784_051B30_EB87', 'S1A_IW_SLC__1SDV_20220418T000312_20220418T000339_042820_051C63_CAB2', 'S1A_IW_SLC__1SDV_20220422T120547_20220422T120614_042886_051E96_41EE', 'S1A_IW_SLC__1SDV_20220427T121354_20220427T121421_042959_0520EE_A42D', 'S1A_IW_SLC__1SDV_20220430T000313_20220430T000340_042995_05221E_46A1', 'S1A_IW_SLC__1SDV_20220504T120547_20220504T120614_043061_05245F_6F62', 'S1A_IW_SLC__1SDV_20220509T121355_20220509T121422_043134_0526C4_092E', 'S1A_IW_SLC__1SDV_20220516T120548_20220516T120615_043236_0529DD_42D2', 'S1A_IW_SLC__1SDV_20220521T121356_20220521T121423_043309_052C00_FE88'], dtype='<U67') - x(x)float646.194e+05 6.195e+05 ... 6.281e+05
- axis :
- X
- long_name :
- x coordinate of projection
- standard_name :
- projection_x_coordinate
- units :
- metre
array([619425., 619455., 619485., ..., 628035., 628065., 628095.])
- y(y)float643.102e+06 3.102e+06 ... 3.09e+06
- axis :
- Y
- long_name :
- y coordinate of projection
- standard_name :
- projection_y_coordinate
- units :
- metre
array([3101655., 3101625., 3101595., ..., 3089865., 3089835., 3089805.])
- spatial_ref()int640
- crs_wkt :
- PROJCS["WGS 84 / UTM zone 45N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",87],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32645"]]
- semi_major_axis :
- 6378137.0
- semi_minor_axis :
- 6356752.314245179
- inverse_flattening :
- 298.257223563
- reference_ellipsoid_name :
- WGS 84
- longitude_of_prime_meridian :
- 0.0
- prime_meridian_name :
- Greenwich
- geographic_crs_name :
- WGS 84
- horizontal_datum_name :
- World Geodetic System 1984
- projected_crs_name :
- WGS 84 / UTM zone 45N
- grid_mapping_name :
- transverse_mercator
- latitude_of_projection_origin :
- 0.0
- longitude_of_central_meridian :
- 87.0
- false_easting :
- 500000.0
- false_northing :
- 0.0
- scale_factor_at_central_meridian :
- 0.9996
- spatial_ref :
- PROJCS["WGS 84 / UTM zone 45N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",87],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32645"]]
- GeoTransform :
- 619410.0 30.0 0.0 3101670.0 0.0 -30.0
array(0)
- sensor(acq_date)<U3'S1A' 'S1A' 'S1A' ... 'S1A' 'S1A'
array(['S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A'], dtype='<U3') - beam_mode(acq_date)<U2'IW' 'IW' 'IW' ... 'IW' 'IW' 'IW'
array(['IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW'], dtype='<U2') - acquisition_time(acq_date)datetime64[ns]2022-10-21T00:03:14 ... 2022-10-...
array(['2022-10-21T00:03:14.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T12:05:45.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:14:20.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:14:14.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:14:20.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T12:05:43.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:05:43.000000000', '2022-10-21T00:03:14.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:52.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T12:13:50.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T12:14:17.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T12:13:54.000000000', ... '2022-10-21T00:03:07.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:50.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T00:03:14.000000000', '2022-10-21T12:05:44.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T00:03:10.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:13:49.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:45.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T00:03:16.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T12:13:51.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T00:03:09.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T00:03:12.000000000'], dtype='datetime64[ns]') - polarisation_type(acq_date)<U2'DV' 'DV' 'DV' ... 'DV' 'DV' 'DV'
array(['DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV'], dtype='<U2') - orbit_type(acq_date)<U1'P' 'P' 'P' 'P' ... 'P' 'P' 'P' 'P'
array(['P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P'], dtype='<U1') - terrain_correction_pixel_spacing(acq_date)<U5'RTC30' 'RTC30' ... 'RTC30' 'RTC30'
array(['RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30'], dtype='<U5') - output_format(acq_date)<U1'g' 'g' 'g' 'g' ... 'g' 'g' 'g' 'g'
array(['g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g'], dtype='<U1') - output_type(acq_date)<U1'p' 'p' 'p' 'p' ... 'p' 'p' 'p' 'p'
array(['p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p'], dtype='<U1') - masked(acq_date)<U1'u' 'u' 'u' 'u' ... 'u' 'u' 'u' 'u'
array(['u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u'], dtype='<U1') - filtered(acq_date)<U1'n' 'n' 'n' 'n' ... 'n' 'n' 'n' 'n'
array(['n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n'], dtype='<U1') - area(acq_date)<U1'e' 'e' 'e' 'e' ... 'e' 'e' 'e' 'e'
array(['e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e'], dtype='<U1') - product_id(acq_date)<U4'748F' '0D1E' ... '5BAD' 'BD36'
array(['748F', '0D1E', '3E89', '61B6', 'F306', '1380', 'ED8C', '2CBE', '1424', 'ABA0', 'E61E', 'E5B6', '5569', '71C9', 'B942', 'DCE1', '6581', '1DFD', '01B2', '9849', 'CFFC', '57F2', 'D4E6', 'B7B0', '3770', '52BB', '0579', 'AA68', '24B8', 'D049', '5FF4', '5F4E', '199D', '0031', 'D082', 'E74B', 'F7ED', '65E0', 'F587', '528F', '51E7', 'F5AA', 'AC38', '772E', 'F0A6', '066E', '5844', '8CD8', '6340', 'D35A', '056C', '6E6F', '0CAD', '6270', '8A4F', 'BEAB', '54B1', '8F7B', 'F9A9', 'AA59', '57E1', '81EB', '250E', '3C39', '3A2A', '9331', 'C14C', 'F1DE', '106F', 'D48F', '407D', 'FD00', '5D96', '971C', 'F573', 'C890', '68EA', '83D7', 'E113', 'D846', 'F4B3', 'BB26', '485C', '6683', '6153', '832B', '7418', '378D', '33F5', '1139', 'B661', '9A2F', '6943', '5BAD', 'BD36'], dtype='<U4') - acq_hour(acq_date)int640 12 12 12 12 12 ... 12 12 0 0 0 0
array([ 0, 12, 12, 12, 12, 12, 0, 0, 12, 12, 0, 12, 0, 12, 0, 12, 0, 12, 0, 12, 12, 12, 0, 0, 12, 0, 12, 12, 12, 12, 12, 0, 0, 12, 0, 12, 12, 12, 12, 12, 12, 12, 0, 12, 12, 12, 12, 0, 12, 0, 12, 12, 12, 12, 12, 12, 0, 12, 12, 0, 12, 12, 12, 12, 12, 0, 12, 12, 0, 0, 12, 12, 12, 12, 12, 12, 0, 12, 12, 0, 0, 0, 12, 12, 12, 12, 12, 12, 12, 12, 12, 0, 0, 0, 0]) - orbital_dir(acq_date)<U4'asc' 'desc' 'desc' ... 'asc' 'asc'
array(['asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'asc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'asc', 'asc'], dtype='<U4') - data_take_id(acq_date)<U6'047321' '047463' ... '052C00'
- ID of data take from SAR acquisition :
- ID data take
array(['047321', '047463', '047676', '047898', '0479A9', '047BBD', '047DE5', '047EF4', '0480FD', '048318', '04841E', '04862F', '04884C', '04895A', '048B6C', '048D87', '048E99', '0490AD', '0493DB', '0495EC', '04980F', '04991E', '049B1F', '049D70', '049EAC', '04A0FF', '04A383', '04A4B9', '04A6FC', '04A972', '04AAB3', '04AD0D', '04AF85', '04B0C4', '04B2FB', '04B566', '04B6AB', '04B8FF', '04BB88', '04BCB8', '04BF12', '04C195', '04C2D6', '04C52E', '04C7A8', '04C8E9', '04CB46', '04CDC8', '04CEFF', '04D14F', '04D3CC', '04D50E', '04D761', '04D9E8', '04DB21', '04DD71', '04DFDD', '04E110', '04E340', '04E5AB', '04E6D7', '04E931', '04EB9B', '04ECCD', '04EEE8', '04F156', '04F294', '04F4D6', '04F754', '04F88B', '04FAF5', '04FD6F', '04FEB2', '050108', '050376', '0504AF', '0506F7', '050962', '050AA2', '050CE5', '050F59', '051096', '0512D7', '05154A', '051682', '0518C0', '051B30', '051C63', '051E96', '0520EE', '05221E', '05245F', '0526C4', '0529DD', '052C00'], dtype='<U6')
- vv(acq_date, y, x)float32dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
- scale_factor :
- 1.0
- add_offset :
- 0.0
- _FillValue :
- 0.0
Array Chunk Bytes 41.62 MiB 448.59 kiB Shape (95, 396, 290) (1, 396, 290) Count 16 Graph Layers 95 Chunks Type float32 numpy.ndarray - vh(acq_date, y, x)float32dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
- scale_factor :
- 1.0
- add_offset :
- 0.0
- _FillValue :
- 0.0
Array Chunk Bytes 41.62 MiB 448.59 kiB Shape (95, 396, 290) (1, 396, 290) Count 24 Graph Layers 95 Chunks Type float32 numpy.ndarray - ls(acq_date, y, x)float64dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
- scale_factor :
- 1.0
- add_offset :
- 0.0
- _FillValue :
- 0
Array Chunk Bytes 83.24 MiB 897.19 kiB Shape (95, 396, 290) (1, 396, 290) Count 28 Graph Layers 95 Chunks Type float64 numpy.ndarray
Before we go further, the nodata value for the ASF dataset is currently zero. Change all zero values to NaN:
vrt_new = vrt_new.where(vrt_new.vv != 0., np.nan, drop=False)
da_pc
<xarray.DataArray 'stackstac-4d29467097b7d384d1232c9847300ccd' (time: 100,
band: 2,
y: 1188, x: 869)>
dask.array<fetch_raster_window, shape=(100, 2, 1188, 869), dtype=float64, chunksize=(1, 1, 1024, 869), chunktype=numpy.ndarray>
Coordinates: (12/40)
* time (time) datetime64[ns] 2021-06-02T1...
id (time) <U66 'S1A_IW_GRDH_1SDV_2021...
* band (band) <U2 'vh' 'vv'
* x (x) float64 6.194e+05 ... 6.281e+05
* y (y) float64 3.102e+06 ... 3.09e+06
sar:looks_azimuth int64 1
... ...
s1:processing_level <U1 '1'
title (band) <U41 'VH: vertical transmit...
description (band) <U173 'Terrain-corrected ga...
raster:bands object {'nodata': -32768, 'data_ty...
epsg int64 32645
granule_id (time) <U62 'S1A_IW_GRDH_1SDV_2021...
Attributes:
spec: RasterSpec(epsg=32645, bounds=(619420.0, 3089780.0, 628110.0...
crs: epsg:32645
transform: | 10.00, 0.00, 619420.00|\n| 0.00,-10.00, 3101660.00|\n| 0.0...
resolution: 10.0- time: 100
- band: 2
- y: 1188
- x: 869
- dask.array<chunksize=(1, 1, 1024, 869), meta=np.ndarray>
Array Chunk Bytes 1.54 GiB 6.79 MiB Shape (100, 2, 1188, 869) (1, 1, 1024, 869) Count 3 Graph Layers 400 Chunks Type float64 numpy.ndarray - time(time)datetime64[ns]2021-06-02T12:05:57.441074 ... 2...
array(['2021-06-02T12:05:57.441074000', '2021-06-07T12:14:05.058343000', '2021-06-10T00:03:23.870450000', '2021-06-14T12:05:58.373830000', '2021-06-19T12:14:05.865662000', '2021-06-22T00:03:24.594621000', '2021-06-26T12:05:58.982444000', '2021-07-01T12:14:06.572473000', '2021-07-04T00:03:25.347747000', '2021-07-08T12:05:59.613554000', '2021-07-13T12:14:07.266919000', '2021-07-16T00:03:26.093195000', '2021-07-20T12:06:00.428473000', '2021-07-25T12:14:07.814919000', '2021-07-28T00:03:26.667023000', '2021-08-01T12:06:01.150857000', '2021-08-06T12:14:08.698596000', '2021-08-09T00:03:27.454588000', '2021-08-13T12:06:01.713793000', '2021-08-18T12:14:09.226637000', '2021-08-21T00:03:28.059716000', '2021-08-25T12:06:02.421490000', '2021-09-02T00:03:28.464707000', '2021-09-06T12:06:02.971474000', '2021-09-11T12:14:10.406593000', '2021-09-14T00:03:29.135941000', '2021-09-18T12:06:03.415938000', '2021-09-23T12:14:10.788780000', '2021-09-26T00:03:29.558327000', '2021-09-30T12:06:03.760613000', '2021-10-08T00:03:29.713275000', '2021-10-12T12:06:03.850150000', '2021-10-17T12:14:11.200359000', '2021-10-20T00:03:29.919736000', '2021-10-24T12:06:03.954256000', '2021-10-29T12:14:11.052769000', '2021-11-01T00:03:29.702246000', '2021-11-05T12:06:03.645002000', '2021-11-10T12:14:10.742983000', '2021-11-13T00:03:29.481869000', '2021-11-17T12:06:03.560526000', '2021-11-22T12:14:10.576899000', '2021-11-25T00:03:29.120003000', '2021-11-29T12:06:03.035677000', '2021-12-04T12:14:10.007158000', '2021-12-07T00:03:28.568419000', '2021-12-11T12:06:02.513966000', '2021-12-16T12:14:09.596874000', '2021-12-19T00:03:28.141449000', '2021-12-23T12:06:01.793721000', '2021-12-28T12:14:08.858303000', '2021-12-31T00:03:27.432317000', '2022-01-04T12:06:01.280484000', '2022-01-09T12:14:08.415805000', '2022-01-12T00:03:27.070182000', '2022-01-16T12:06:00.877035000', '2022-01-21T12:14:07.716165000', '2022-01-24T00:03:26.424832000', '2022-01-28T12:06:00.352862000', '2022-02-02T12:14:07.057390000', '2022-02-14T12:14:07.178103000', '2022-02-17T00:03:25.677942000', '2022-02-21T12:05:59.939806000', '2022-02-26T12:14:07.075742000', '2022-03-01T00:03:25.589986000', '2022-03-05T12:05:59.677073000', '2022-03-10T12:14:06.994606000', '2022-03-13T00:03:25.744897000', '2022-03-17T12:05:59.706124000', '2022-03-22T12:14:07.362180000', '2022-03-25T00:03:26.139523000', '2022-03-29T12:06:00.140262000', '2022-04-03T12:14:07.585924000', '2022-04-06T00:03:26.181999000', '2022-04-10T12:06:00.398442000', '2022-04-15T12:14:07.711367000', '2022-04-18T00:03:26.603587000', '2022-04-22T12:06:01.051309000', '2022-04-27T12:14:08.426000000', '2022-04-30T00:03:27.201903000', '2022-05-04T12:06:01.122366000', '2022-05-09T12:14:08.980398000', '2022-05-16T12:06:02.314833000', '2022-05-21T12:14:09.786375000', '2022-05-28T12:06:03.110968000', '2022-06-02T12:14:10.897104000', '2022-06-05T00:03:29.836741000', '2022-06-09T12:06:04.189105000', '2022-06-14T12:14:11.513880000', '2022-06-17T00:03:30.273763000', '2022-06-21T12:06:04.879869000', '2022-06-26T12:14:12.455112000', '2022-07-03T12:06:05.553402000', '2022-07-08T12:14:13.035532000', '2022-07-11T00:03:31.868452000', '2022-07-15T12:06:06.242032000', '2022-07-20T12:14:13.801000000', '2022-07-23T00:03:32.650978000', '2022-07-27T12:06:06.953202000', '2022-08-01T12:14:14.610123000'], dtype='datetime64[ns]') - id(time)<U66'S1A_IW_GRDH_1SDV_20210602T12054...
array(['S1A_IW_GRDH_1SDV_20210602T120544_20210602T120609_038161_0480FD_rtc', 'S1A_IW_GRDH_1SDV_20210607T121352_20210607T121417_038234_048318_rtc', 'S1A_IW_GRDH_1SDV_20210610T000311_20210610T000336_038270_04841E_rtc', 'S1A_IW_GRDH_1SDV_20210614T120545_20210614T120610_038336_04862F_rtc', 'S1A_IW_GRDH_1SDV_20210619T121353_20210619T121418_038409_04884C_rtc', 'S1A_IW_GRDH_1SDV_20210622T000312_20210622T000337_038445_04895A_rtc', 'S1A_IW_GRDH_1SDV_20210626T120546_20210626T120611_038511_048B6C_rtc', 'S1A_IW_GRDH_1SDV_20210701T121354_20210701T121419_038584_048D87_rtc', 'S1A_IW_GRDH_1SDV_20210704T000312_20210704T000337_038620_048E99_rtc', 'S1A_IW_GRDH_1SDV_20210708T120547_20210708T120612_038686_0490AD_rtc', 'S1A_IW_GRDH_1SDV_20210713T121354_20210713T121419_038759_0492D4_rtc', 'S1A_IW_GRDH_1SDV_20210716T000313_20210716T000338_038795_0493DB_rtc', 'S1A_IW_GRDH_1SDV_20210720T120547_20210720T120612_038861_0495EC_rtc', 'S1A_IW_GRDH_1SDV_20210725T121355_20210725T121420_038934_04980F_rtc', 'S1A_IW_GRDH_1SDV_20210728T000314_20210728T000339_038970_04991E_rtc', 'S1A_IW_GRDH_1SDV_20210801T120548_20210801T120613_039036_049B1F_rtc', 'S1A_IW_GRDH_1SDV_20210806T121356_20210806T121421_039109_049D70_rtc', 'S1A_IW_GRDH_1SDV_20210809T000314_20210809T000339_039145_049EAC_rtc', 'S1A_IW_GRDH_1SDV_20210813T120549_20210813T120614_039211_04A0FF_rtc', 'S1A_IW_GRDH_1SDV_20210818T121356_20210818T121421_039284_04A383_rtc', ... 'S1A_IW_GRDH_1SDV_20220509T121356_20220509T121421_043134_0526C4_rtc', 'S1A_IW_GRDH_1SDV_20220516T120549_20220516T120614_043236_0529DD_rtc', 'S1A_IW_GRDH_1SDV_20220521T121357_20220521T121422_043309_052C00_rtc', 'S1A_IW_GRDH_1SDV_20220528T120550_20220528T120615_043411_052F0A_rtc', 'S1A_IW_GRDH_1SDV_20220602T121358_20220602T121423_043484_053126_rtc', 'S1A_IW_GRDH_1SDV_20220605T000317_20220605T000342_043520_05323C_rtc', 'S1A_IW_GRDH_1SDV_20220609T120551_20220609T120616_043586_053436_rtc', 'S1A_IW_GRDH_1SDV_20220614T121359_20220614T121424_043659_05365F_rtc', 'S1A_IW_GRDH_1SDV_20220617T000317_20220617T000342_043695_053770_rtc', 'S1A_IW_GRDH_1SDV_20220621T120552_20220621T120617_043761_053978_rtc', 'S1A_IW_GRDH_1SDV_20220626T121359_20220626T121424_043834_053BA2_rtc', 'S1A_IW_GRDH_1SDV_20220703T120553_20220703T120618_043936_053EA7_rtc', 'S1A_IW_GRDH_1SDV_20220708T121400_20220708T121425_044009_0540D0_rtc', 'S1A_IW_GRDH_1SDV_20220711T000319_20220711T000344_044045_0541E4_rtc', 'S1A_IW_GRDH_1SDV_20220715T120553_20220715T120618_044111_0543E4_rtc', 'S1A_IW_GRDH_1SDV_20220720T121401_20220720T121426_044184_054615_rtc', 'S1A_IW_GRDH_1SDV_20220723T000320_20220723T000345_044220_05471E_rtc', 'S1A_IW_GRDH_1SDV_20220727T120554_20220727T120619_044286_05491D_rtc', 'S1A_IW_GRDH_1SDV_20220801T121402_20220801T121427_044359_054B39_rtc'], dtype='<U66') - band(band)<U2'vh' 'vv'
array(['vh', 'vv'], dtype='<U2')
- x(x)float646.194e+05 6.194e+05 ... 6.281e+05
array([619420., 619430., 619440., ..., 628080., 628090., 628100.])
- y(y)float643.102e+06 3.102e+06 ... 3.09e+06
array([3101660., 3101650., 3101640., ..., 3089810., 3089800., 3089790.])
- sar:looks_azimuth()int641
array(1)
- sat:relative_orbit(time)int64114 12 48 114 12 ... 12 48 114 12
array([114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 114, 12, 114, 12, 48, 114, 12, 48, 114, 12, 114, 12, 48, 114, 12, 48, 114, 12]) - s1:instrument_configuration_ID(time)<U1'6' '6' '6' '6' ... '7' '7' '7' '7'
array(['6', '6', '6', '6', '6', '6', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7'], dtype='<U1') - s1:orbit_source(time)<U8'DOWNLINK' 'DOWNLINK' ... 'RESORB'
array(['DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB'], dtype='<U8') - end_datetime(time)<U32'2021-06-02 12:06:09.940813+00:0...
array(['2021-06-02 12:06:09.940813+00:00', '2021-06-07 12:14:17.557268+00:00', '2021-06-10 00:03:36.369821+00:00', '2021-06-14 12:06:10.872859+00:00', '2021-06-19 12:14:18.365410+00:00', '2021-06-22 00:03:37.093898+00:00', '2021-06-26 12:06:11.482149+00:00', '2021-07-01 12:14:19.071405+00:00', '2021-07-04 00:03:37.847082+00:00', '2021-07-08 12:06:12.112559+00:00', '2021-07-13 12:14:19.766644+00:00', '2021-07-16 00:03:38.592472+00:00', '2021-07-20 12:06:12.927516+00:00', '2021-07-25 12:14:20.313830+00:00', '2021-07-28 00:03:39.166376+00:00', '2021-08-01 12:06:13.649807+00:00', '2021-08-06 12:14:21.197519+00:00', '2021-08-09 00:03:39.953924+00:00', '2021-08-13 12:06:14.213502+00:00', '2021-08-18 12:14:21.725577+00:00', ... '2022-05-04 12:06:13.622093+00:00', '2022-05-09 12:14:21.479341+00:00', '2022-05-16 12:06:14.814536+00:00', '2022-05-21 12:14:22.285304+00:00', '2022-05-28 12:06:15.610707+00:00', '2022-06-02 12:14:23.396823+00:00', '2022-06-05 00:03:42.336084+00:00', '2022-06-09 12:06:16.688822+00:00', '2022-06-14 12:14:24.012807+00:00', '2022-06-17 00:03:42.773155+00:00', '2022-06-21 12:06:17.379612+00:00', '2022-06-26 12:14:24.954817+00:00', '2022-07-03 12:06:18.052440+00:00', '2022-07-08 12:14:25.534456+00:00', '2022-07-11 00:03:44.367804+00:00', '2022-07-15 12:06:18.741751+00:00', '2022-07-20 12:14:26.299940+00:00', '2022-07-23 00:03:45.150333+00:00', '2022-07-27 12:06:19.452211+00:00', '2022-08-01 12:14:27.109058+00:00'], dtype='<U32') - sat:orbit_state(time)<U10'ascending' ... 'ascending'
array(['ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'ascending', 'ascending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending'], dtype='<U10') - platform()<U11'SENTINEL-1A'
array('SENTINEL-1A', dtype='<U11') - s1:slice_number(time)<U2'6' '6' '12' '6' ... '12' '6' '6'
array(['6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '6', '6', '6', '6', '12', '6', '6', '12', '6', '6', '6', '6', '12', '6', '6', '12', '6', '6'], dtype='<U2') - start_datetime(time)<U32'2021-06-02 12:05:44.941334+00:0...
array(['2021-06-02 12:05:44.941334+00:00', '2021-06-07 12:13:52.559418+00:00', '2021-06-10 00:03:11.371078+00:00', '2021-06-14 12:05:45.874802+00:00', '2021-06-19 12:13:53.365914+00:00', '2021-06-22 00:03:12.095344+00:00', '2021-06-26 12:05:46.482738+00:00', '2021-07-01 12:13:54.073541+00:00', '2021-07-04 00:03:12.848412+00:00', '2021-07-08 12:05:47.114550+00:00', '2021-07-13 12:13:54.767193+00:00', '2021-07-16 00:03:13.593918+00:00', '2021-07-20 12:05:47.929430+00:00', '2021-07-25 12:13:55.316007+00:00', '2021-07-28 00:03:14.167671+00:00', '2021-08-01 12:05:48.651907+00:00', '2021-08-06 12:13:56.199672+00:00', '2021-08-09 00:03:14.955252+00:00', '2021-08-13 12:05:49.214084+00:00', '2021-08-18 12:13:56.727697+00:00', ... '2022-05-04 12:05:48.622638+00:00', '2022-05-09 12:13:56.481455+00:00', '2022-05-16 12:05:49.815129+00:00', '2022-05-21 12:13:57.287447+00:00', '2022-05-28 12:05:50.611230+00:00', '2022-06-02 12:13:58.397386+00:00', '2022-06-05 00:03:17.337399+00:00', '2022-06-09 12:05:51.689387+00:00', '2022-06-14 12:13:59.014953+00:00', '2022-06-17 00:03:17.774371+00:00', '2022-06-21 12:05:52.380127+00:00', '2022-06-26 12:13:59.955407+00:00', '2022-07-03 12:05:53.054364+00:00', '2022-07-08 12:14:00.536608+00:00', '2022-07-11 00:03:19.369100+00:00', '2022-07-15 12:05:53.742314+00:00', '2022-07-20 12:14:01.302060+00:00', '2022-07-23 00:03:20.151622+00:00', '2022-07-27 12:05:54.454193+00:00', '2022-08-01 12:14:02.111187+00:00'], dtype='<U32') - sar:resolution_azimuth()int6422
array(22)
- s1:resolution()<U4'high'
array('high', dtype='<U4') - sar:frequency_band()<U1'C'
array('C', dtype='<U1') - sar:center_frequency()float645.405
array(5.405)
- sar:pixel_spacing_azimuth()int6410
array(10)
- sat:platform_international_designator()<U9'2014-016A'
array('2014-016A', dtype='<U9') - constellation()<U10'Sentinel-1'
array('Sentinel-1', dtype='<U10') - sar:polarizations()object{'VV', 'VH'}
array({'VV', 'VH'}, dtype=object) - proj:epsg()int6432645
array(32645)
- sar:pixel_spacing_range()int6410
array(10)
- sar:looks_range()int645
array(5)
- sar:resolution_range()int6420
array(20)
- sat:absolute_orbit(time)int6438161 38234 38270 ... 44286 44359
array([38161, 38234, 38270, 38336, 38409, 38445, 38511, 38584, 38620, 38686, 38759, 38795, 38861, 38934, 38970, 39036, 39109, 39145, 39211, 39284, 39320, 39386, 39495, 39561, 39634, 39670, 39736, 39809, 39845, 39911, 40020, 40086, 40159, 40195, 40261, 40334, 40370, 40436, 40509, 40545, 40611, 40684, 40720, 40786, 40859, 40895, 40961, 41034, 41070, 41136, 41209, 41245, 41311, 41384, 41420, 41486, 41559, 41595, 41661, 41734, 41909, 41945, 42011, 42084, 42120, 42186, 42259, 42295, 42361, 42434, 42470, 42536, 42609, 42645, 42711, 42784, 42820, 42886, 42959, 42995, 43061, 43134, 43236, 43309, 43411, 43484, 43520, 43586, 43659, 43695, 43761, 43834, 43936, 44009, 44045, 44111, 44184, 44220, 44286, 44359]) - sar:looks_equivalent_number()float644.4
array(4.4)
- sar:observation_direction()<U5'right'
array('right', dtype='<U5') - sar:product_type()<U3'GRD'
array('GRD', dtype='<U3') - s1:product_timeliness()<U8'Fast-24h'
array('Fast-24h', dtype='<U8') - sar:instrument_mode()<U2'IW'
array('IW', dtype='<U2') - s1:datatake_id(time)<U6'295165' '295704' ... '346937'
array(['295165', '295704', '295966', '296495', '297036', '297306', '297836', '298375', '298649', '299181', '299732', '299995', '300524', '301071', '301342', '301855', '302448', '302764', '303359', '304003', '304313', '304892', '305843', '306445', '307077', '307396', '307963', '308582', '308907', '309503', '310456', '311058', '311701', '312022', '312622', '313256', '313577', '314182', '314824', '315135', '315727', '316364', '316686', '317281', '317928', '318241', '318833', '319453', '319760', '320320', '320939', '321239', '321841', '322459', '322765', '323304', '323926', '324244', '324822', '325460', '327023', '327346', '327944', '328566', '328879', '329463', '330082', '330402', '330981', '331609', '331926', '332503', '333130', '333442', '334016', '334640', '334947', '335510', '336110', '336414', '336991', '337604', '338397', '338944', '339722', '340262', '340540', '341046', '341599', '341872', '342392', '342946', '343719', '344272', '344548', '345060', '345621', '345886', '346397', '346937'], dtype='<U6') - s1:total_slices(time)<U2'21' '20' '17' ... '17' '21' '20'
array(['21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '21', '20', '21', '20', '17', '21', '20', '17', '21', '20', '21', '20', '17', '21', '20', '17', '21', '20'], dtype='<U2') - s1:processing_level()<U1'1'
array('1', dtype='<U1') - title(band)<U41'VH: vertical transmit, horizont...
array(['VH: vertical transmit, horizontal receive', 'VV: vertical transmit, vertical receive'], dtype='<U41') - description(band)<U173'Terrain-corrected gamma naught ...
array(['Terrain-corrected gamma naught values of signal transmitted with vertical polarization and received with horizontal polarization with radiometric terrain correction applied.', 'Terrain-corrected gamma naught values of signal transmitted with vertical polarization and received with vertical polarization with radiometric terrain correction applied.'], dtype='<U173') - raster:bands()object{'nodata': -32768, 'data_type': ...
array({'nodata': -32768, 'data_type': 'float32', 'spatial_resolution': 10.0}, dtype=object) - epsg()int6432645
array(32645)
- granule_id(time)<U62'S1A_IW_GRDH_1SDV_20210602T12054...
array(['S1A_IW_GRDH_1SDV_20210602T120544_20210602T120609_038161_0480FD', 'S1A_IW_GRDH_1SDV_20210607T121352_20210607T121417_038234_048318', 'S1A_IW_GRDH_1SDV_20210610T000311_20210610T000336_038270_04841E', 'S1A_IW_GRDH_1SDV_20210614T120545_20210614T120610_038336_04862F', 'S1A_IW_GRDH_1SDV_20210619T121353_20210619T121418_038409_04884C', 'S1A_IW_GRDH_1SDV_20210622T000312_20210622T000337_038445_04895A', 'S1A_IW_GRDH_1SDV_20210626T120546_20210626T120611_038511_048B6C', 'S1A_IW_GRDH_1SDV_20210701T121354_20210701T121419_038584_048D87', 'S1A_IW_GRDH_1SDV_20210704T000312_20210704T000337_038620_048E99', 'S1A_IW_GRDH_1SDV_20210708T120547_20210708T120612_038686_0490AD', 'S1A_IW_GRDH_1SDV_20210713T121354_20210713T121419_038759_0492D4', 'S1A_IW_GRDH_1SDV_20210716T000313_20210716T000338_038795_0493DB', 'S1A_IW_GRDH_1SDV_20210720T120547_20210720T120612_038861_0495EC', 'S1A_IW_GRDH_1SDV_20210725T121355_20210725T121420_038934_04980F', 'S1A_IW_GRDH_1SDV_20210728T000314_20210728T000339_038970_04991E', 'S1A_IW_GRDH_1SDV_20210801T120548_20210801T120613_039036_049B1F', 'S1A_IW_GRDH_1SDV_20210806T121356_20210806T121421_039109_049D70', 'S1A_IW_GRDH_1SDV_20210809T000314_20210809T000339_039145_049EAC', 'S1A_IW_GRDH_1SDV_20210813T120549_20210813T120614_039211_04A0FF', 'S1A_IW_GRDH_1SDV_20210818T121356_20210818T121421_039284_04A383', ... 'S1A_IW_GRDH_1SDV_20220509T121356_20220509T121421_043134_0526C4', 'S1A_IW_GRDH_1SDV_20220516T120549_20220516T120614_043236_0529DD', 'S1A_IW_GRDH_1SDV_20220521T121357_20220521T121422_043309_052C00', 'S1A_IW_GRDH_1SDV_20220528T120550_20220528T120615_043411_052F0A', 'S1A_IW_GRDH_1SDV_20220602T121358_20220602T121423_043484_053126', 'S1A_IW_GRDH_1SDV_20220605T000317_20220605T000342_043520_05323C', 'S1A_IW_GRDH_1SDV_20220609T120551_20220609T120616_043586_053436', 'S1A_IW_GRDH_1SDV_20220614T121359_20220614T121424_043659_05365F', 'S1A_IW_GRDH_1SDV_20220617T000317_20220617T000342_043695_053770', 'S1A_IW_GRDH_1SDV_20220621T120552_20220621T120617_043761_053978', 'S1A_IW_GRDH_1SDV_20220626T121359_20220626T121424_043834_053BA2', 'S1A_IW_GRDH_1SDV_20220703T120553_20220703T120618_043936_053EA7', 'S1A_IW_GRDH_1SDV_20220708T121400_20220708T121425_044009_0540D0', 'S1A_IW_GRDH_1SDV_20220711T000319_20220711T000344_044045_0541E4', 'S1A_IW_GRDH_1SDV_20220715T120553_20220715T120618_044111_0543E4', 'S1A_IW_GRDH_1SDV_20220720T121401_20220720T121426_044184_054615', 'S1A_IW_GRDH_1SDV_20220723T000320_20220723T000345_044220_05471E', 'S1A_IW_GRDH_1SDV_20220727T120554_20220727T120619_044286_05491D', 'S1A_IW_GRDH_1SDV_20220801T121402_20220801T121427_044359_054B39'], dtype='<U62')
- spec :
- RasterSpec(epsg=32645, bounds=(619420.0, 3089780.0, 628110.0, 3101660.0), resolutions_xy=(10.0, 10.0))
- crs :
- epsg:32645
- transform :
- | 10.00, 0.00, 619420.00| | 0.00,-10.00, 3101660.00| | 0.00, 0.00, 1.00|
- resolution :
- 10.0
Extract common data take ID from granule IDs#
We want to ensure that we are performing a direct comparison of the ASF and PC datasets. To do this, we would like to use the acquisition ID that is stored in the source granule name (published by ESA). In the setup notebooks we attached the entire granule IDs of the SLC images to the ASF dataset and the GRD images to the PC dataset. In the ASF data inspection notebook, we attached data take id as a non-dimensional coordinate. Now we will do the same for the Planetary Computer dataset, extracting just the 6-digit acquisition ID from the granule ID and using this for a scene-by-scene comparison.
data_take_pc = [str(da_pc.isel(time=t).granule_id.values)[56:] for t in range(len(da_pc.time))]
Assign data_take_id as a non-dimensional coordinate. Rather than use the xr.assign_coords() function, we are adding the object by specifying a tuple with the form (‘coord_name’, coord_data, attrs):
da_pc.coords['data_take_id'] = ('time', data_take_pc)
vrt_new
<xarray.Dataset>
Dimensions: (acq_date: 95, y: 396, x: 290)
Coordinates: (12/20)
* acq_date (acq_date) datetime64[ns] 2021-05-02 .....
granule_id (acq_date) <U67 'S1A_IW_SLC__1SDV_20210...
* x (x) float64 6.194e+05 ... 6.281e+05
* y (y) float64 3.102e+06 ... 3.09e+06
spatial_ref int64 0
sensor (acq_date) <U3 'S1A' 'S1A' ... 'S1A' 'S1A'
... ...
filtered (acq_date) <U1 'n' 'n' 'n' ... 'n' 'n' 'n'
area (acq_date) <U1 'e' 'e' 'e' ... 'e' 'e' 'e'
product_id (acq_date) <U4 '748F' '0D1E' ... 'BD36'
acq_hour (acq_date) int64 0 12 12 12 12 ... 0 0 0 0
orbital_dir (acq_date) <U4 'asc' 'desc' ... 'asc'
data_take_id (acq_date) <U6 '047321' ... '052C00'
Data variables:
vv (acq_date, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
vh (acq_date, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
ls (acq_date, y, x) float64 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>- acq_date: 95
- y: 396
- x: 290
- acq_date(acq_date)datetime64[ns]2021-05-02 ... 2022-05-21
array(['2021-05-02T00:00:00.000000000', '2021-05-05T00:00:00.000000000', '2021-05-09T00:00:00.000000000', '2021-05-14T00:00:00.000000000', '2021-05-17T00:00:00.000000000', '2021-05-21T00:00:00.000000000', '2021-05-26T00:00:00.000000000', '2021-05-29T00:00:00.000000000', '2021-06-02T00:00:00.000000000', '2021-06-07T00:00:00.000000000', '2021-06-10T00:00:00.000000000', '2021-06-14T00:00:00.000000000', '2021-06-19T00:00:00.000000000', '2021-06-22T00:00:00.000000000', '2021-06-26T00:00:00.000000000', '2021-07-01T00:00:00.000000000', '2021-07-04T00:00:00.000000000', '2021-07-08T00:00:00.000000000', '2021-07-16T00:00:00.000000000', '2021-07-20T00:00:00.000000000', '2021-07-25T00:00:00.000000000', '2021-07-28T00:00:00.000000000', '2021-08-01T00:00:00.000000000', '2021-08-06T00:00:00.000000000', '2021-08-09T00:00:00.000000000', '2021-08-13T00:00:00.000000000', '2021-08-18T00:00:00.000000000', '2021-08-21T00:00:00.000000000', '2021-08-25T00:00:00.000000000', '2021-08-30T00:00:00.000000000', '2021-09-02T00:00:00.000000000', '2021-09-06T00:00:00.000000000', '2021-09-11T00:00:00.000000000', '2021-09-14T00:00:00.000000000', '2021-09-18T00:00:00.000000000', '2021-09-23T00:00:00.000000000', '2021-09-26T00:00:00.000000000', '2021-09-30T00:00:00.000000000', '2021-10-05T00:00:00.000000000', '2021-10-08T00:00:00.000000000', '2021-10-12T00:00:00.000000000', '2021-10-17T00:00:00.000000000', '2021-10-20T00:00:00.000000000', '2021-10-24T00:00:00.000000000', '2021-10-29T00:00:00.000000000', '2021-11-01T00:00:00.000000000', '2021-11-05T00:00:00.000000000', '2021-11-10T00:00:00.000000000', '2021-11-13T00:00:00.000000000', '2021-11-17T00:00:00.000000000', '2021-11-22T00:00:00.000000000', '2021-11-25T00:00:00.000000000', '2021-11-29T00:00:00.000000000', '2021-12-04T00:00:00.000000000', '2021-12-07T00:00:00.000000000', '2021-12-11T00:00:00.000000000', '2021-12-16T00:00:00.000000000', '2021-12-19T00:00:00.000000000', '2021-12-23T00:00:00.000000000', '2021-12-28T00:00:00.000000000', '2021-12-31T00:00:00.000000000', '2022-01-04T00:00:00.000000000', '2022-01-09T00:00:00.000000000', '2022-01-12T00:00:00.000000000', '2022-01-16T00:00:00.000000000', '2022-01-21T00:00:00.000000000', '2022-01-24T00:00:00.000000000', '2022-01-28T00:00:00.000000000', '2022-02-02T00:00:00.000000000', '2022-02-05T00:00:00.000000000', '2022-02-09T00:00:00.000000000', '2022-02-14T00:00:00.000000000', '2022-02-17T00:00:00.000000000', '2022-02-21T00:00:00.000000000', '2022-02-26T00:00:00.000000000', '2022-03-01T00:00:00.000000000', '2022-03-05T00:00:00.000000000', '2022-03-10T00:00:00.000000000', '2022-03-13T00:00:00.000000000', '2022-03-17T00:00:00.000000000', '2022-03-22T00:00:00.000000000', '2022-03-25T00:00:00.000000000', '2022-03-29T00:00:00.000000000', '2022-04-03T00:00:00.000000000', '2022-04-06T00:00:00.000000000', '2022-04-10T00:00:00.000000000', '2022-04-15T00:00:00.000000000', '2022-04-18T00:00:00.000000000', '2022-04-22T00:00:00.000000000', '2022-04-27T00:00:00.000000000', '2022-04-30T00:00:00.000000000', '2022-05-04T00:00:00.000000000', '2022-05-09T00:00:00.000000000', '2022-05-16T00:00:00.000000000', '2022-05-21T00:00:00.000000000'], dtype='datetime64[ns]') - granule_id(acq_date)<U67'S1A_IW_SLC__1SDV_20210502T12141...
- description :
- source granule ID for ASF-processed S1 RTC imagery, extracted from README files for each scene
array(['S1A_IW_SLC__1SDV_20210502T121414_20210502T121441_037709_047321_900F', 'S1A_IW_SLC__1SDV_20210505T000307_20210505T000334_037745_047463_4DEF', 'S1A_IW_SLC__1SDV_20210509T120542_20210509T120609_037811_047676_9EBD', 'S1A_IW_SLC__1SDV_20210514T121349_20210514T121416_037884_047898_4702', 'S1A_IW_SLC__1SDV_20210517T000308_20210517T000335_037920_0479A9_2A18', 'S1A_IW_SLC__1SDV_20210521T120543_20210521T120609_037986_047BBD_1CA0', 'S1A_IW_SLC__1SDV_20210526T121350_20210526T121417_038059_047DE5_0E67', 'S1A_IW_SLC__1SDV_20210529T000309_20210529T000336_038095_047EF4_5449', 'S1A_IW_SLC__1SDV_20210602T120543_20210602T120610_038161_0480FD_893F', 'S1A_IW_SLC__1SDV_20210607T121351_20210607T121418_038234_048318_9B64', 'S1A_IW_SLC__1SDV_20210610T000310_20210610T000337_038270_04841E_601A', 'S1A_IW_SLC__1SDV_20210614T120544_20210614T120611_038336_04862F_337B', 'S1A_IW_SLC__1SDV_20210619T121352_20210619T121419_038409_04884C_1DBA', 'S1A_IW_SLC__1SDV_20210622T000310_20210622T000337_038445_04895A_842E', 'S1A_IW_SLC__1SDV_20210626T120545_20210626T120612_038511_048B6C_9585', 'S1A_IW_SLC__1SDV_20210701T121352_20210701T121419_038584_048D87_F95C', 'S1A_IW_SLC__1SDV_20210704T000311_20210704T000338_038620_048E99_1BAA', 'S1A_IW_SLC__1SDV_20210708T120545_20210708T120612_038686_0490AD_DD9E', 'S1A_IW_SLC__1SDV_20210716T000312_20210716T000339_038795_0493DB_53CD', 'S1A_IW_SLC__1SDV_20210720T120546_20210720T120613_038861_0495EC_C9D4', ... 'S1A_IW_SLC__1SDV_20220305T120545_20220305T120612_042186_0506F7_1601', 'S1A_IW_SLC__1SDV_20220310T121353_20220310T121420_042259_050962_068B', 'S1A_IW_SLC__1SDV_20220313T000311_20220313T000338_042295_050AA2_2909', 'S1A_IW_SLC__1SDV_20220317T120545_20220317T120612_042361_050CE5_4CCF', 'S1A_IW_SLC__1SDV_20220322T121353_20220322T121420_042434_050F59_A1B6', 'S1A_IW_SLC__1SDV_20220325T000312_20220325T000339_042470_051096_BA6F', 'S1A_IW_SLC__1SDV_20220329T120546_20220329T120613_042536_0512D7_425A', 'S1A_IW_SLC__1SDV_20220403T121353_20220403T121420_042609_05154A_3FE7', 'S1A_IW_SLC__1SDV_20220406T000312_20220406T000339_042645_051682_4B40', 'S1A_IW_SLC__1SDV_20220410T120546_20220410T120613_042711_0518C0_3349', 'S1A_IW_SLC__1SDV_20220415T121353_20220415T121420_042784_051B30_EB87', 'S1A_IW_SLC__1SDV_20220418T000312_20220418T000339_042820_051C63_CAB2', 'S1A_IW_SLC__1SDV_20220422T120547_20220422T120614_042886_051E96_41EE', 'S1A_IW_SLC__1SDV_20220427T121354_20220427T121421_042959_0520EE_A42D', 'S1A_IW_SLC__1SDV_20220430T000313_20220430T000340_042995_05221E_46A1', 'S1A_IW_SLC__1SDV_20220504T120547_20220504T120614_043061_05245F_6F62', 'S1A_IW_SLC__1SDV_20220509T121355_20220509T121422_043134_0526C4_092E', 'S1A_IW_SLC__1SDV_20220516T120548_20220516T120615_043236_0529DD_42D2', 'S1A_IW_SLC__1SDV_20220521T121356_20220521T121423_043309_052C00_FE88'], dtype='<U67') - x(x)float646.194e+05 6.195e+05 ... 6.281e+05
- axis :
- X
- long_name :
- x coordinate of projection
- standard_name :
- projection_x_coordinate
- units :
- metre
array([619425., 619455., 619485., ..., 628035., 628065., 628095.])
- y(y)float643.102e+06 3.102e+06 ... 3.09e+06
- axis :
- Y
- long_name :
- y coordinate of projection
- standard_name :
- projection_y_coordinate
- units :
- metre
array([3101655., 3101625., 3101595., ..., 3089865., 3089835., 3089805.])
- spatial_ref()int640
- crs_wkt :
- PROJCS["WGS 84 / UTM zone 45N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",87],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32645"]]
- semi_major_axis :
- 6378137.0
- semi_minor_axis :
- 6356752.314245179
- inverse_flattening :
- 298.257223563
- reference_ellipsoid_name :
- WGS 84
- longitude_of_prime_meridian :
- 0.0
- prime_meridian_name :
- Greenwich
- geographic_crs_name :
- WGS 84
- horizontal_datum_name :
- World Geodetic System 1984
- projected_crs_name :
- WGS 84 / UTM zone 45N
- grid_mapping_name :
- transverse_mercator
- latitude_of_projection_origin :
- 0.0
- longitude_of_central_meridian :
- 87.0
- false_easting :
- 500000.0
- false_northing :
- 0.0
- scale_factor_at_central_meridian :
- 0.9996
- spatial_ref :
- PROJCS["WGS 84 / UTM zone 45N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",87],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32645"]]
- GeoTransform :
- 619410.0 30.0 0.0 3101670.0 0.0 -30.0
array(0)
- sensor(acq_date)<U3'S1A' 'S1A' 'S1A' ... 'S1A' 'S1A'
array(['S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A'], dtype='<U3') - beam_mode(acq_date)<U2'IW' 'IW' 'IW' ... 'IW' 'IW' 'IW'
array(['IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW'], dtype='<U2') - acquisition_time(acq_date)datetime64[ns]2022-10-21T00:03:14 ... 2022-10-...
array(['2022-10-21T00:03:14.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T12:05:45.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:14:20.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:14:14.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:14:20.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T12:05:43.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:05:43.000000000', '2022-10-21T00:03:14.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:52.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T12:13:50.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T12:14:17.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T12:13:54.000000000', ... '2022-10-21T00:03:07.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:50.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T00:03:14.000000000', '2022-10-21T12:05:44.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T00:03:10.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:13:49.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:45.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T00:03:16.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T12:13:51.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T00:03:09.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T00:03:12.000000000'], dtype='datetime64[ns]') - polarisation_type(acq_date)<U2'DV' 'DV' 'DV' ... 'DV' 'DV' 'DV'
array(['DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV'], dtype='<U2') - orbit_type(acq_date)<U1'P' 'P' 'P' 'P' ... 'P' 'P' 'P' 'P'
array(['P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P'], dtype='<U1') - terrain_correction_pixel_spacing(acq_date)<U5'RTC30' 'RTC30' ... 'RTC30' 'RTC30'
array(['RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30'], dtype='<U5') - output_format(acq_date)<U1'g' 'g' 'g' 'g' ... 'g' 'g' 'g' 'g'
array(['g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g'], dtype='<U1') - output_type(acq_date)<U1'p' 'p' 'p' 'p' ... 'p' 'p' 'p' 'p'
array(['p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p'], dtype='<U1') - masked(acq_date)<U1'u' 'u' 'u' 'u' ... 'u' 'u' 'u' 'u'
array(['u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u'], dtype='<U1') - filtered(acq_date)<U1'n' 'n' 'n' 'n' ... 'n' 'n' 'n' 'n'
array(['n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n'], dtype='<U1') - area(acq_date)<U1'e' 'e' 'e' 'e' ... 'e' 'e' 'e' 'e'
array(['e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e'], dtype='<U1') - product_id(acq_date)<U4'748F' '0D1E' ... '5BAD' 'BD36'
array(['748F', '0D1E', '3E89', '61B6', 'F306', '1380', 'ED8C', '2CBE', '1424', 'ABA0', 'E61E', 'E5B6', '5569', '71C9', 'B942', 'DCE1', '6581', '1DFD', '01B2', '9849', 'CFFC', '57F2', 'D4E6', 'B7B0', '3770', '52BB', '0579', 'AA68', '24B8', 'D049', '5FF4', '5F4E', '199D', '0031', 'D082', 'E74B', 'F7ED', '65E0', 'F587', '528F', '51E7', 'F5AA', 'AC38', '772E', 'F0A6', '066E', '5844', '8CD8', '6340', 'D35A', '056C', '6E6F', '0CAD', '6270', '8A4F', 'BEAB', '54B1', '8F7B', 'F9A9', 'AA59', '57E1', '81EB', '250E', '3C39', '3A2A', '9331', 'C14C', 'F1DE', '106F', 'D48F', '407D', 'FD00', '5D96', '971C', 'F573', 'C890', '68EA', '83D7', 'E113', 'D846', 'F4B3', 'BB26', '485C', '6683', '6153', '832B', '7418', '378D', '33F5', '1139', 'B661', '9A2F', '6943', '5BAD', 'BD36'], dtype='<U4') - acq_hour(acq_date)int640 12 12 12 12 12 ... 12 12 0 0 0 0
array([ 0, 12, 12, 12, 12, 12, 0, 0, 12, 12, 0, 12, 0, 12, 0, 12, 0, 12, 0, 12, 12, 12, 0, 0, 12, 0, 12, 12, 12, 12, 12, 0, 0, 12, 0, 12, 12, 12, 12, 12, 12, 12, 0, 12, 12, 12, 12, 0, 12, 0, 12, 12, 12, 12, 12, 12, 0, 12, 12, 0, 12, 12, 12, 12, 12, 0, 12, 12, 0, 0, 12, 12, 12, 12, 12, 12, 0, 12, 12, 0, 0, 0, 12, 12, 12, 12, 12, 12, 12, 12, 12, 0, 0, 0, 0]) - orbital_dir(acq_date)<U4'asc' 'desc' 'desc' ... 'asc' 'asc'
array(['asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'asc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'asc', 'asc'], dtype='<U4') - data_take_id(acq_date)<U6'047321' '047463' ... '052C00'
- ID of data take from SAR acquisition :
- ID data take
array(['047321', '047463', '047676', '047898', '0479A9', '047BBD', '047DE5', '047EF4', '0480FD', '048318', '04841E', '04862F', '04884C', '04895A', '048B6C', '048D87', '048E99', '0490AD', '0493DB', '0495EC', '04980F', '04991E', '049B1F', '049D70', '049EAC', '04A0FF', '04A383', '04A4B9', '04A6FC', '04A972', '04AAB3', '04AD0D', '04AF85', '04B0C4', '04B2FB', '04B566', '04B6AB', '04B8FF', '04BB88', '04BCB8', '04BF12', '04C195', '04C2D6', '04C52E', '04C7A8', '04C8E9', '04CB46', '04CDC8', '04CEFF', '04D14F', '04D3CC', '04D50E', '04D761', '04D9E8', '04DB21', '04DD71', '04DFDD', '04E110', '04E340', '04E5AB', '04E6D7', '04E931', '04EB9B', '04ECCD', '04EEE8', '04F156', '04F294', '04F4D6', '04F754', '04F88B', '04FAF5', '04FD6F', '04FEB2', '050108', '050376', '0504AF', '0506F7', '050962', '050AA2', '050CE5', '050F59', '051096', '0512D7', '05154A', '051682', '0518C0', '051B30', '051C63', '051E96', '0520EE', '05221E', '05245F', '0526C4', '0529DD', '052C00'], dtype='<U6')
- vv(acq_date, y, x)float32dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
- scale_factor :
- 1.0
- add_offset :
- 0.0
- _FillValue :
- 0.0
Array Chunk Bytes 41.62 MiB 448.59 kiB Shape (95, 396, 290) (1, 396, 290) Count 18 Graph Layers 95 Chunks Type float32 numpy.ndarray - vh(acq_date, y, x)float32dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
- scale_factor :
- 1.0
- add_offset :
- 0.0
- _FillValue :
- 0.0
Array Chunk Bytes 41.62 MiB 448.59 kiB Shape (95, 396, 290) (1, 396, 290) Count 29 Graph Layers 95 Chunks Type float32 numpy.ndarray - ls(acq_date, y, x)float64dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
- scale_factor :
- 1.0
- add_offset :
- 0.0
- _FillValue :
- 0
Array Chunk Bytes 83.24 MiB 897.19 kiB Shape (95, 396, 290) (1, 396, 290) Count 33 Graph Layers 95 Chunks Type float64 numpy.ndarray
da_pc
<xarray.DataArray 'stackstac-4d29467097b7d384d1232c9847300ccd' (time: 100,
band: 2,
y: 1188, x: 869)>
dask.array<fetch_raster_window, shape=(100, 2, 1188, 869), dtype=float64, chunksize=(1, 1, 1024, 869), chunktype=numpy.ndarray>
Coordinates: (12/41)
* time (time) datetime64[ns] 2021-06-02T1...
id (time) <U66 'S1A_IW_GRDH_1SDV_2021...
* band (band) <U2 'vh' 'vv'
* x (x) float64 6.194e+05 ... 6.281e+05
* y (y) float64 3.102e+06 ... 3.09e+06
sar:looks_azimuth int64 1
... ...
title (band) <U41 'VH: vertical transmit...
description (band) <U173 'Terrain-corrected ga...
raster:bands object {'nodata': -32768, 'data_ty...
epsg int64 32645
granule_id (time) <U62 'S1A_IW_GRDH_1SDV_2021...
data_take_id (time) <U6 '0480FD' ... '054B39'
Attributes:
spec: RasterSpec(epsg=32645, bounds=(619420.0, 3089780.0, 628110.0...
crs: epsg:32645
transform: | 10.00, 0.00, 619420.00|\n| 0.00,-10.00, 3101660.00|\n| 0.0...
resolution: 10.0- time: 100
- band: 2
- y: 1188
- x: 869
- dask.array<chunksize=(1, 1, 1024, 869), meta=np.ndarray>
Array Chunk Bytes 1.54 GiB 6.79 MiB Shape (100, 2, 1188, 869) (1, 1, 1024, 869) Count 3 Graph Layers 400 Chunks Type float64 numpy.ndarray - time(time)datetime64[ns]2021-06-02T12:05:57.441074 ... 2...
array(['2021-06-02T12:05:57.441074000', '2021-06-07T12:14:05.058343000', '2021-06-10T00:03:23.870450000', '2021-06-14T12:05:58.373830000', '2021-06-19T12:14:05.865662000', '2021-06-22T00:03:24.594621000', '2021-06-26T12:05:58.982444000', '2021-07-01T12:14:06.572473000', '2021-07-04T00:03:25.347747000', '2021-07-08T12:05:59.613554000', '2021-07-13T12:14:07.266919000', '2021-07-16T00:03:26.093195000', '2021-07-20T12:06:00.428473000', '2021-07-25T12:14:07.814919000', '2021-07-28T00:03:26.667023000', '2021-08-01T12:06:01.150857000', '2021-08-06T12:14:08.698596000', '2021-08-09T00:03:27.454588000', '2021-08-13T12:06:01.713793000', '2021-08-18T12:14:09.226637000', '2021-08-21T00:03:28.059716000', '2021-08-25T12:06:02.421490000', '2021-09-02T00:03:28.464707000', '2021-09-06T12:06:02.971474000', '2021-09-11T12:14:10.406593000', '2021-09-14T00:03:29.135941000', '2021-09-18T12:06:03.415938000', '2021-09-23T12:14:10.788780000', '2021-09-26T00:03:29.558327000', '2021-09-30T12:06:03.760613000', '2021-10-08T00:03:29.713275000', '2021-10-12T12:06:03.850150000', '2021-10-17T12:14:11.200359000', '2021-10-20T00:03:29.919736000', '2021-10-24T12:06:03.954256000', '2021-10-29T12:14:11.052769000', '2021-11-01T00:03:29.702246000', '2021-11-05T12:06:03.645002000', '2021-11-10T12:14:10.742983000', '2021-11-13T00:03:29.481869000', '2021-11-17T12:06:03.560526000', '2021-11-22T12:14:10.576899000', '2021-11-25T00:03:29.120003000', '2021-11-29T12:06:03.035677000', '2021-12-04T12:14:10.007158000', '2021-12-07T00:03:28.568419000', '2021-12-11T12:06:02.513966000', '2021-12-16T12:14:09.596874000', '2021-12-19T00:03:28.141449000', '2021-12-23T12:06:01.793721000', '2021-12-28T12:14:08.858303000', '2021-12-31T00:03:27.432317000', '2022-01-04T12:06:01.280484000', '2022-01-09T12:14:08.415805000', '2022-01-12T00:03:27.070182000', '2022-01-16T12:06:00.877035000', '2022-01-21T12:14:07.716165000', '2022-01-24T00:03:26.424832000', '2022-01-28T12:06:00.352862000', '2022-02-02T12:14:07.057390000', '2022-02-14T12:14:07.178103000', '2022-02-17T00:03:25.677942000', '2022-02-21T12:05:59.939806000', '2022-02-26T12:14:07.075742000', '2022-03-01T00:03:25.589986000', '2022-03-05T12:05:59.677073000', '2022-03-10T12:14:06.994606000', '2022-03-13T00:03:25.744897000', '2022-03-17T12:05:59.706124000', '2022-03-22T12:14:07.362180000', '2022-03-25T00:03:26.139523000', '2022-03-29T12:06:00.140262000', '2022-04-03T12:14:07.585924000', '2022-04-06T00:03:26.181999000', '2022-04-10T12:06:00.398442000', '2022-04-15T12:14:07.711367000', '2022-04-18T00:03:26.603587000', '2022-04-22T12:06:01.051309000', '2022-04-27T12:14:08.426000000', '2022-04-30T00:03:27.201903000', '2022-05-04T12:06:01.122366000', '2022-05-09T12:14:08.980398000', '2022-05-16T12:06:02.314833000', '2022-05-21T12:14:09.786375000', '2022-05-28T12:06:03.110968000', '2022-06-02T12:14:10.897104000', '2022-06-05T00:03:29.836741000', '2022-06-09T12:06:04.189105000', '2022-06-14T12:14:11.513880000', '2022-06-17T00:03:30.273763000', '2022-06-21T12:06:04.879869000', '2022-06-26T12:14:12.455112000', '2022-07-03T12:06:05.553402000', '2022-07-08T12:14:13.035532000', '2022-07-11T00:03:31.868452000', '2022-07-15T12:06:06.242032000', '2022-07-20T12:14:13.801000000', '2022-07-23T00:03:32.650978000', '2022-07-27T12:06:06.953202000', '2022-08-01T12:14:14.610123000'], dtype='datetime64[ns]') - id(time)<U66'S1A_IW_GRDH_1SDV_20210602T12054...
array(['S1A_IW_GRDH_1SDV_20210602T120544_20210602T120609_038161_0480FD_rtc', 'S1A_IW_GRDH_1SDV_20210607T121352_20210607T121417_038234_048318_rtc', 'S1A_IW_GRDH_1SDV_20210610T000311_20210610T000336_038270_04841E_rtc', 'S1A_IW_GRDH_1SDV_20210614T120545_20210614T120610_038336_04862F_rtc', 'S1A_IW_GRDH_1SDV_20210619T121353_20210619T121418_038409_04884C_rtc', 'S1A_IW_GRDH_1SDV_20210622T000312_20210622T000337_038445_04895A_rtc', 'S1A_IW_GRDH_1SDV_20210626T120546_20210626T120611_038511_048B6C_rtc', 'S1A_IW_GRDH_1SDV_20210701T121354_20210701T121419_038584_048D87_rtc', 'S1A_IW_GRDH_1SDV_20210704T000312_20210704T000337_038620_048E99_rtc', 'S1A_IW_GRDH_1SDV_20210708T120547_20210708T120612_038686_0490AD_rtc', 'S1A_IW_GRDH_1SDV_20210713T121354_20210713T121419_038759_0492D4_rtc', 'S1A_IW_GRDH_1SDV_20210716T000313_20210716T000338_038795_0493DB_rtc', 'S1A_IW_GRDH_1SDV_20210720T120547_20210720T120612_038861_0495EC_rtc', 'S1A_IW_GRDH_1SDV_20210725T121355_20210725T121420_038934_04980F_rtc', 'S1A_IW_GRDH_1SDV_20210728T000314_20210728T000339_038970_04991E_rtc', 'S1A_IW_GRDH_1SDV_20210801T120548_20210801T120613_039036_049B1F_rtc', 'S1A_IW_GRDH_1SDV_20210806T121356_20210806T121421_039109_049D70_rtc', 'S1A_IW_GRDH_1SDV_20210809T000314_20210809T000339_039145_049EAC_rtc', 'S1A_IW_GRDH_1SDV_20210813T120549_20210813T120614_039211_04A0FF_rtc', 'S1A_IW_GRDH_1SDV_20210818T121356_20210818T121421_039284_04A383_rtc', ... 'S1A_IW_GRDH_1SDV_20220509T121356_20220509T121421_043134_0526C4_rtc', 'S1A_IW_GRDH_1SDV_20220516T120549_20220516T120614_043236_0529DD_rtc', 'S1A_IW_GRDH_1SDV_20220521T121357_20220521T121422_043309_052C00_rtc', 'S1A_IW_GRDH_1SDV_20220528T120550_20220528T120615_043411_052F0A_rtc', 'S1A_IW_GRDH_1SDV_20220602T121358_20220602T121423_043484_053126_rtc', 'S1A_IW_GRDH_1SDV_20220605T000317_20220605T000342_043520_05323C_rtc', 'S1A_IW_GRDH_1SDV_20220609T120551_20220609T120616_043586_053436_rtc', 'S1A_IW_GRDH_1SDV_20220614T121359_20220614T121424_043659_05365F_rtc', 'S1A_IW_GRDH_1SDV_20220617T000317_20220617T000342_043695_053770_rtc', 'S1A_IW_GRDH_1SDV_20220621T120552_20220621T120617_043761_053978_rtc', 'S1A_IW_GRDH_1SDV_20220626T121359_20220626T121424_043834_053BA2_rtc', 'S1A_IW_GRDH_1SDV_20220703T120553_20220703T120618_043936_053EA7_rtc', 'S1A_IW_GRDH_1SDV_20220708T121400_20220708T121425_044009_0540D0_rtc', 'S1A_IW_GRDH_1SDV_20220711T000319_20220711T000344_044045_0541E4_rtc', 'S1A_IW_GRDH_1SDV_20220715T120553_20220715T120618_044111_0543E4_rtc', 'S1A_IW_GRDH_1SDV_20220720T121401_20220720T121426_044184_054615_rtc', 'S1A_IW_GRDH_1SDV_20220723T000320_20220723T000345_044220_05471E_rtc', 'S1A_IW_GRDH_1SDV_20220727T120554_20220727T120619_044286_05491D_rtc', 'S1A_IW_GRDH_1SDV_20220801T121402_20220801T121427_044359_054B39_rtc'], dtype='<U66') - band(band)<U2'vh' 'vv'
array(['vh', 'vv'], dtype='<U2')
- x(x)float646.194e+05 6.194e+05 ... 6.281e+05
array([619420., 619430., 619440., ..., 628080., 628090., 628100.])
- y(y)float643.102e+06 3.102e+06 ... 3.09e+06
array([3101660., 3101650., 3101640., ..., 3089810., 3089800., 3089790.])
- sar:looks_azimuth()int641
array(1)
- sat:relative_orbit(time)int64114 12 48 114 12 ... 12 48 114 12
array([114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 114, 12, 114, 12, 48, 114, 12, 48, 114, 12, 114, 12, 48, 114, 12, 48, 114, 12]) - s1:instrument_configuration_ID(time)<U1'6' '6' '6' '6' ... '7' '7' '7' '7'
array(['6', '6', '6', '6', '6', '6', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7'], dtype='<U1') - s1:orbit_source(time)<U8'DOWNLINK' 'DOWNLINK' ... 'RESORB'
array(['DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB'], dtype='<U8') - end_datetime(time)<U32'2021-06-02 12:06:09.940813+00:0...
array(['2021-06-02 12:06:09.940813+00:00', '2021-06-07 12:14:17.557268+00:00', '2021-06-10 00:03:36.369821+00:00', '2021-06-14 12:06:10.872859+00:00', '2021-06-19 12:14:18.365410+00:00', '2021-06-22 00:03:37.093898+00:00', '2021-06-26 12:06:11.482149+00:00', '2021-07-01 12:14:19.071405+00:00', '2021-07-04 00:03:37.847082+00:00', '2021-07-08 12:06:12.112559+00:00', '2021-07-13 12:14:19.766644+00:00', '2021-07-16 00:03:38.592472+00:00', '2021-07-20 12:06:12.927516+00:00', '2021-07-25 12:14:20.313830+00:00', '2021-07-28 00:03:39.166376+00:00', '2021-08-01 12:06:13.649807+00:00', '2021-08-06 12:14:21.197519+00:00', '2021-08-09 00:03:39.953924+00:00', '2021-08-13 12:06:14.213502+00:00', '2021-08-18 12:14:21.725577+00:00', ... '2022-05-04 12:06:13.622093+00:00', '2022-05-09 12:14:21.479341+00:00', '2022-05-16 12:06:14.814536+00:00', '2022-05-21 12:14:22.285304+00:00', '2022-05-28 12:06:15.610707+00:00', '2022-06-02 12:14:23.396823+00:00', '2022-06-05 00:03:42.336084+00:00', '2022-06-09 12:06:16.688822+00:00', '2022-06-14 12:14:24.012807+00:00', '2022-06-17 00:03:42.773155+00:00', '2022-06-21 12:06:17.379612+00:00', '2022-06-26 12:14:24.954817+00:00', '2022-07-03 12:06:18.052440+00:00', '2022-07-08 12:14:25.534456+00:00', '2022-07-11 00:03:44.367804+00:00', '2022-07-15 12:06:18.741751+00:00', '2022-07-20 12:14:26.299940+00:00', '2022-07-23 00:03:45.150333+00:00', '2022-07-27 12:06:19.452211+00:00', '2022-08-01 12:14:27.109058+00:00'], dtype='<U32') - sat:orbit_state(time)<U10'ascending' ... 'ascending'
array(['ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'ascending', 'ascending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending'], dtype='<U10') - platform()<U11'SENTINEL-1A'
array('SENTINEL-1A', dtype='<U11') - s1:slice_number(time)<U2'6' '6' '12' '6' ... '12' '6' '6'
array(['6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '6', '6', '6', '6', '12', '6', '6', '12', '6', '6', '6', '6', '12', '6', '6', '12', '6', '6'], dtype='<U2') - start_datetime(time)<U32'2021-06-02 12:05:44.941334+00:0...
array(['2021-06-02 12:05:44.941334+00:00', '2021-06-07 12:13:52.559418+00:00', '2021-06-10 00:03:11.371078+00:00', '2021-06-14 12:05:45.874802+00:00', '2021-06-19 12:13:53.365914+00:00', '2021-06-22 00:03:12.095344+00:00', '2021-06-26 12:05:46.482738+00:00', '2021-07-01 12:13:54.073541+00:00', '2021-07-04 00:03:12.848412+00:00', '2021-07-08 12:05:47.114550+00:00', '2021-07-13 12:13:54.767193+00:00', '2021-07-16 00:03:13.593918+00:00', '2021-07-20 12:05:47.929430+00:00', '2021-07-25 12:13:55.316007+00:00', '2021-07-28 00:03:14.167671+00:00', '2021-08-01 12:05:48.651907+00:00', '2021-08-06 12:13:56.199672+00:00', '2021-08-09 00:03:14.955252+00:00', '2021-08-13 12:05:49.214084+00:00', '2021-08-18 12:13:56.727697+00:00', ... '2022-05-04 12:05:48.622638+00:00', '2022-05-09 12:13:56.481455+00:00', '2022-05-16 12:05:49.815129+00:00', '2022-05-21 12:13:57.287447+00:00', '2022-05-28 12:05:50.611230+00:00', '2022-06-02 12:13:58.397386+00:00', '2022-06-05 00:03:17.337399+00:00', '2022-06-09 12:05:51.689387+00:00', '2022-06-14 12:13:59.014953+00:00', '2022-06-17 00:03:17.774371+00:00', '2022-06-21 12:05:52.380127+00:00', '2022-06-26 12:13:59.955407+00:00', '2022-07-03 12:05:53.054364+00:00', '2022-07-08 12:14:00.536608+00:00', '2022-07-11 00:03:19.369100+00:00', '2022-07-15 12:05:53.742314+00:00', '2022-07-20 12:14:01.302060+00:00', '2022-07-23 00:03:20.151622+00:00', '2022-07-27 12:05:54.454193+00:00', '2022-08-01 12:14:02.111187+00:00'], dtype='<U32') - sar:resolution_azimuth()int6422
array(22)
- s1:resolution()<U4'high'
array('high', dtype='<U4') - sar:frequency_band()<U1'C'
array('C', dtype='<U1') - sar:center_frequency()float645.405
array(5.405)
- sar:pixel_spacing_azimuth()int6410
array(10)
- sat:platform_international_designator()<U9'2014-016A'
array('2014-016A', dtype='<U9') - constellation()<U10'Sentinel-1'
array('Sentinel-1', dtype='<U10') - sar:polarizations()object{'VV', 'VH'}
array({'VV', 'VH'}, dtype=object) - proj:epsg()int6432645
array(32645)
- sar:pixel_spacing_range()int6410
array(10)
- sar:looks_range()int645
array(5)
- sar:resolution_range()int6420
array(20)
- sat:absolute_orbit(time)int6438161 38234 38270 ... 44286 44359
array([38161, 38234, 38270, 38336, 38409, 38445, 38511, 38584, 38620, 38686, 38759, 38795, 38861, 38934, 38970, 39036, 39109, 39145, 39211, 39284, 39320, 39386, 39495, 39561, 39634, 39670, 39736, 39809, 39845, 39911, 40020, 40086, 40159, 40195, 40261, 40334, 40370, 40436, 40509, 40545, 40611, 40684, 40720, 40786, 40859, 40895, 40961, 41034, 41070, 41136, 41209, 41245, 41311, 41384, 41420, 41486, 41559, 41595, 41661, 41734, 41909, 41945, 42011, 42084, 42120, 42186, 42259, 42295, 42361, 42434, 42470, 42536, 42609, 42645, 42711, 42784, 42820, 42886, 42959, 42995, 43061, 43134, 43236, 43309, 43411, 43484, 43520, 43586, 43659, 43695, 43761, 43834, 43936, 44009, 44045, 44111, 44184, 44220, 44286, 44359]) - sar:looks_equivalent_number()float644.4
array(4.4)
- sar:observation_direction()<U5'right'
array('right', dtype='<U5') - sar:product_type()<U3'GRD'
array('GRD', dtype='<U3') - s1:product_timeliness()<U8'Fast-24h'
array('Fast-24h', dtype='<U8') - sar:instrument_mode()<U2'IW'
array('IW', dtype='<U2') - s1:datatake_id(time)<U6'295165' '295704' ... '346937'
array(['295165', '295704', '295966', '296495', '297036', '297306', '297836', '298375', '298649', '299181', '299732', '299995', '300524', '301071', '301342', '301855', '302448', '302764', '303359', '304003', '304313', '304892', '305843', '306445', '307077', '307396', '307963', '308582', '308907', '309503', '310456', '311058', '311701', '312022', '312622', '313256', '313577', '314182', '314824', '315135', '315727', '316364', '316686', '317281', '317928', '318241', '318833', '319453', '319760', '320320', '320939', '321239', '321841', '322459', '322765', '323304', '323926', '324244', '324822', '325460', '327023', '327346', '327944', '328566', '328879', '329463', '330082', '330402', '330981', '331609', '331926', '332503', '333130', '333442', '334016', '334640', '334947', '335510', '336110', '336414', '336991', '337604', '338397', '338944', '339722', '340262', '340540', '341046', '341599', '341872', '342392', '342946', '343719', '344272', '344548', '345060', '345621', '345886', '346397', '346937'], dtype='<U6') - s1:total_slices(time)<U2'21' '20' '17' ... '17' '21' '20'
array(['21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '21', '20', '21', '20', '17', '21', '20', '17', '21', '20', '21', '20', '17', '21', '20', '17', '21', '20'], dtype='<U2') - s1:processing_level()<U1'1'
array('1', dtype='<U1') - title(band)<U41'VH: vertical transmit, horizont...
array(['VH: vertical transmit, horizontal receive', 'VV: vertical transmit, vertical receive'], dtype='<U41') - description(band)<U173'Terrain-corrected gamma naught ...
array(['Terrain-corrected gamma naught values of signal transmitted with vertical polarization and received with horizontal polarization with radiometric terrain correction applied.', 'Terrain-corrected gamma naught values of signal transmitted with vertical polarization and received with vertical polarization with radiometric terrain correction applied.'], dtype='<U173') - raster:bands()object{'nodata': -32768, 'data_type': ...
array({'nodata': -32768, 'data_type': 'float32', 'spatial_resolution': 10.0}, dtype=object) - epsg()int6432645
array(32645)
- granule_id(time)<U62'S1A_IW_GRDH_1SDV_20210602T12054...
array(['S1A_IW_GRDH_1SDV_20210602T120544_20210602T120609_038161_0480FD', 'S1A_IW_GRDH_1SDV_20210607T121352_20210607T121417_038234_048318', 'S1A_IW_GRDH_1SDV_20210610T000311_20210610T000336_038270_04841E', 'S1A_IW_GRDH_1SDV_20210614T120545_20210614T120610_038336_04862F', 'S1A_IW_GRDH_1SDV_20210619T121353_20210619T121418_038409_04884C', 'S1A_IW_GRDH_1SDV_20210622T000312_20210622T000337_038445_04895A', 'S1A_IW_GRDH_1SDV_20210626T120546_20210626T120611_038511_048B6C', 'S1A_IW_GRDH_1SDV_20210701T121354_20210701T121419_038584_048D87', 'S1A_IW_GRDH_1SDV_20210704T000312_20210704T000337_038620_048E99', 'S1A_IW_GRDH_1SDV_20210708T120547_20210708T120612_038686_0490AD', 'S1A_IW_GRDH_1SDV_20210713T121354_20210713T121419_038759_0492D4', 'S1A_IW_GRDH_1SDV_20210716T000313_20210716T000338_038795_0493DB', 'S1A_IW_GRDH_1SDV_20210720T120547_20210720T120612_038861_0495EC', 'S1A_IW_GRDH_1SDV_20210725T121355_20210725T121420_038934_04980F', 'S1A_IW_GRDH_1SDV_20210728T000314_20210728T000339_038970_04991E', 'S1A_IW_GRDH_1SDV_20210801T120548_20210801T120613_039036_049B1F', 'S1A_IW_GRDH_1SDV_20210806T121356_20210806T121421_039109_049D70', 'S1A_IW_GRDH_1SDV_20210809T000314_20210809T000339_039145_049EAC', 'S1A_IW_GRDH_1SDV_20210813T120549_20210813T120614_039211_04A0FF', 'S1A_IW_GRDH_1SDV_20210818T121356_20210818T121421_039284_04A383', ... 'S1A_IW_GRDH_1SDV_20220509T121356_20220509T121421_043134_0526C4', 'S1A_IW_GRDH_1SDV_20220516T120549_20220516T120614_043236_0529DD', 'S1A_IW_GRDH_1SDV_20220521T121357_20220521T121422_043309_052C00', 'S1A_IW_GRDH_1SDV_20220528T120550_20220528T120615_043411_052F0A', 'S1A_IW_GRDH_1SDV_20220602T121358_20220602T121423_043484_053126', 'S1A_IW_GRDH_1SDV_20220605T000317_20220605T000342_043520_05323C', 'S1A_IW_GRDH_1SDV_20220609T120551_20220609T120616_043586_053436', 'S1A_IW_GRDH_1SDV_20220614T121359_20220614T121424_043659_05365F', 'S1A_IW_GRDH_1SDV_20220617T000317_20220617T000342_043695_053770', 'S1A_IW_GRDH_1SDV_20220621T120552_20220621T120617_043761_053978', 'S1A_IW_GRDH_1SDV_20220626T121359_20220626T121424_043834_053BA2', 'S1A_IW_GRDH_1SDV_20220703T120553_20220703T120618_043936_053EA7', 'S1A_IW_GRDH_1SDV_20220708T121400_20220708T121425_044009_0540D0', 'S1A_IW_GRDH_1SDV_20220711T000319_20220711T000344_044045_0541E4', 'S1A_IW_GRDH_1SDV_20220715T120553_20220715T120618_044111_0543E4', 'S1A_IW_GRDH_1SDV_20220720T121401_20220720T121426_044184_054615', 'S1A_IW_GRDH_1SDV_20220723T000320_20220723T000345_044220_05471E', 'S1A_IW_GRDH_1SDV_20220727T120554_20220727T120619_044286_05491D', 'S1A_IW_GRDH_1SDV_20220801T121402_20220801T121427_044359_054B39'], dtype='<U62') - data_take_id(time)<U6'0480FD' '048318' ... '054B39'
array(['0480FD', '048318', '04841E', '04862F', '04884C', '04895A', '048B6C', '048D87', '048E99', '0490AD', '0492D4', '0493DB', '0495EC', '04980F', '04991E', '049B1F', '049D70', '049EAC', '04A0FF', '04A383', '04A4B9', '04A6FC', '04AAB3', '04AD0D', '04AF85', '04B0C4', '04B2FB', '04B566', '04B6AB', '04B8FF', '04BCB8', '04BF12', '04C195', '04C2D6', '04C52E', '04C7A8', '04C8E9', '04CB46', '04CDC8', '04CEFF', '04D14F', '04D3CC', '04D50E', '04D761', '04D9E8', '04DB21', '04DD71', '04DFDD', '04E110', '04E340', '04E5AB', '04E6D7', '04E931', '04EB9B', '04ECCD', '04EEE8', '04F156', '04F294', '04F4D6', '04F754', '04FD6F', '04FEB2', '050108', '050376', '0504AF', '0506F7', '050962', '050AA2', '050CE5', '050F59', '051096', '0512D7', '05154A', '051682', '0518C0', '051B30', '051C63', '051E96', '0520EE', '05221E', '05245F', '0526C4', '0529DD', '052C00', '052F0A', '053126', '05323C', '053436', '05365F', '053770', '053978', '053BA2', '053EA7', '0540D0', '0541E4', '0543E4', '054615', '05471E', '05491D', '054B39'], dtype='<U6')
- spec :
- RasterSpec(epsg=32645, bounds=(619420.0, 3089780.0, 628110.0, 3101660.0), resolutions_xy=(10.0, 10.0))
- crs :
- epsg:32645
- transform :
- | 10.00, 0.00, 619420.00| | 0.00,-10.00, 3101660.00| | 0.00, 0.00, 1.00|
- resolution :
- 10.0
Subset to only common data takes#
Now we have data take ID coordinates for both datasets. We want to find the common data take IDs between the two datasets. To do this, I extract a list of the acquisition IDs (data_take_id) for both datasets and then find the intersection of the two lists (the list object common_data_takes)
pc_data_take_ls = list(da_pc.data_take_id.values)
asf_data_take_ls = list(vrt_new.data_take_id.values)
common_data_takes = list(set(pc_data_take_ls) & set(asf_data_take_ls))
len(common_data_takes)
83
It looks like there are 83 RTC images that are generated from common acquisitions between the two datasets
We’d like to subset the ASF and the PC datasets to only the common acquisitions. The xarray .isin() method is very useful for this type of selection.
subset_condition_asf = vrt_new.data_take_id.isin(common_data_takes)
subset_condition_pc = da_pc.data_take_id.isin(common_data_takes)
Next, we want to select the elements along the acq_ate (and time) dimensions where data_take_id satisfies the requirement of being in the common_data_takes list. I found this stack overflow answer useful for this step: https://stackoverflow.com/questions/70777676/xarray-select-dataarray-according-to-an-non-dimension-coordinate.
asf_subset = vrt_new.sel(acq_date = vrt_new.data_take_id.isin(common_data_takes))
pc_subset = da_pc.sel(time = da_pc.data_take_id.isin(common_data_takes))
pc_subset
<xarray.DataArray 'stackstac-4d29467097b7d384d1232c9847300ccd' (time: 83,
band: 2,
y: 1188, x: 869)>
dask.array<getitem, shape=(83, 2, 1188, 869), dtype=float64, chunksize=(1, 1, 1024, 869), chunktype=numpy.ndarray>
Coordinates: (12/41)
* time (time) datetime64[ns] 2021-06-02T1...
id (time) <U66 'S1A_IW_GRDH_1SDV_2021...
* band (band) <U2 'vh' 'vv'
* x (x) float64 6.194e+05 ... 6.281e+05
* y (y) float64 3.102e+06 ... 3.09e+06
sar:looks_azimuth int64 1
... ...
title (band) <U41 'VH: vertical transmit...
description (band) <U173 'Terrain-corrected ga...
raster:bands object {'nodata': -32768, 'data_ty...
epsg int64 32645
granule_id (time) <U62 'S1A_IW_GRDH_1SDV_2021...
data_take_id (time) <U6 '0480FD' ... '052C00'
Attributes:
spec: RasterSpec(epsg=32645, bounds=(619420.0, 3089780.0, 628110.0...
crs: epsg:32645
transform: | 10.00, 0.00, 619420.00|\n| 0.00,-10.00, 3101660.00|\n| 0.0...
resolution: 10.0- time: 83
- band: 2
- y: 1188
- x: 869
- dask.array<chunksize=(1, 1, 1024, 869), meta=np.ndarray>
Array Chunk Bytes 1.28 GiB 6.79 MiB Shape (83, 2, 1188, 869) (1, 1, 1024, 869) Count 4 Graph Layers 332 Chunks Type float64 numpy.ndarray - time(time)datetime64[ns]2021-06-02T12:05:57.441074 ... 2...
array(['2021-06-02T12:05:57.441074000', '2021-06-07T12:14:05.058343000', '2021-06-10T00:03:23.870450000', '2021-06-14T12:05:58.373830000', '2021-06-19T12:14:05.865662000', '2021-06-22T00:03:24.594621000', '2021-06-26T12:05:58.982444000', '2021-07-01T12:14:06.572473000', '2021-07-04T00:03:25.347747000', '2021-07-08T12:05:59.613554000', '2021-07-16T00:03:26.093195000', '2021-07-20T12:06:00.428473000', '2021-07-25T12:14:07.814919000', '2021-07-28T00:03:26.667023000', '2021-08-01T12:06:01.150857000', '2021-08-06T12:14:08.698596000', '2021-08-09T00:03:27.454588000', '2021-08-13T12:06:01.713793000', '2021-08-18T12:14:09.226637000', '2021-08-21T00:03:28.059716000', '2021-08-25T12:06:02.421490000', '2021-09-02T00:03:28.464707000', '2021-09-06T12:06:02.971474000', '2021-09-11T12:14:10.406593000', '2021-09-14T00:03:29.135941000', '2021-09-18T12:06:03.415938000', '2021-09-23T12:14:10.788780000', '2021-09-26T00:03:29.558327000', '2021-09-30T12:06:03.760613000', '2021-10-08T00:03:29.713275000', '2021-10-12T12:06:03.850150000', '2021-10-17T12:14:11.200359000', '2021-10-20T00:03:29.919736000', '2021-10-24T12:06:03.954256000', '2021-10-29T12:14:11.052769000', '2021-11-01T00:03:29.702246000', '2021-11-05T12:06:03.645002000', '2021-11-10T12:14:10.742983000', '2021-11-13T00:03:29.481869000', '2021-11-17T12:06:03.560526000', '2021-11-22T12:14:10.576899000', '2021-11-25T00:03:29.120003000', '2021-11-29T12:06:03.035677000', '2021-12-04T12:14:10.007158000', '2021-12-07T00:03:28.568419000', '2021-12-11T12:06:02.513966000', '2021-12-16T12:14:09.596874000', '2021-12-19T00:03:28.141449000', '2021-12-23T12:06:01.793721000', '2021-12-28T12:14:08.858303000', '2021-12-31T00:03:27.432317000', '2022-01-04T12:06:01.280484000', '2022-01-09T12:14:08.415805000', '2022-01-12T00:03:27.070182000', '2022-01-16T12:06:00.877035000', '2022-01-21T12:14:07.716165000', '2022-01-24T00:03:26.424832000', '2022-01-28T12:06:00.352862000', '2022-02-02T12:14:07.057390000', '2022-02-14T12:14:07.178103000', '2022-02-17T00:03:25.677942000', '2022-02-21T12:05:59.939806000', '2022-02-26T12:14:07.075742000', '2022-03-01T00:03:25.589986000', '2022-03-05T12:05:59.677073000', '2022-03-10T12:14:06.994606000', '2022-03-13T00:03:25.744897000', '2022-03-17T12:05:59.706124000', '2022-03-22T12:14:07.362180000', '2022-03-25T00:03:26.139523000', '2022-03-29T12:06:00.140262000', '2022-04-03T12:14:07.585924000', '2022-04-06T00:03:26.181999000', '2022-04-10T12:06:00.398442000', '2022-04-15T12:14:07.711367000', '2022-04-18T00:03:26.603587000', '2022-04-22T12:06:01.051309000', '2022-04-27T12:14:08.426000000', '2022-04-30T00:03:27.201903000', '2022-05-04T12:06:01.122366000', '2022-05-09T12:14:08.980398000', '2022-05-16T12:06:02.314833000', '2022-05-21T12:14:09.786375000'], dtype='datetime64[ns]') - id(time)<U66'S1A_IW_GRDH_1SDV_20210602T12054...
array(['S1A_IW_GRDH_1SDV_20210602T120544_20210602T120609_038161_0480FD_rtc', 'S1A_IW_GRDH_1SDV_20210607T121352_20210607T121417_038234_048318_rtc', 'S1A_IW_GRDH_1SDV_20210610T000311_20210610T000336_038270_04841E_rtc', 'S1A_IW_GRDH_1SDV_20210614T120545_20210614T120610_038336_04862F_rtc', 'S1A_IW_GRDH_1SDV_20210619T121353_20210619T121418_038409_04884C_rtc', 'S1A_IW_GRDH_1SDV_20210622T000312_20210622T000337_038445_04895A_rtc', 'S1A_IW_GRDH_1SDV_20210626T120546_20210626T120611_038511_048B6C_rtc', 'S1A_IW_GRDH_1SDV_20210701T121354_20210701T121419_038584_048D87_rtc', 'S1A_IW_GRDH_1SDV_20210704T000312_20210704T000337_038620_048E99_rtc', 'S1A_IW_GRDH_1SDV_20210708T120547_20210708T120612_038686_0490AD_rtc', 'S1A_IW_GRDH_1SDV_20210716T000313_20210716T000338_038795_0493DB_rtc', 'S1A_IW_GRDH_1SDV_20210720T120547_20210720T120612_038861_0495EC_rtc', 'S1A_IW_GRDH_1SDV_20210725T121355_20210725T121420_038934_04980F_rtc', 'S1A_IW_GRDH_1SDV_20210728T000314_20210728T000339_038970_04991E_rtc', 'S1A_IW_GRDH_1SDV_20210801T120548_20210801T120613_039036_049B1F_rtc', 'S1A_IW_GRDH_1SDV_20210806T121356_20210806T121421_039109_049D70_rtc', 'S1A_IW_GRDH_1SDV_20210809T000314_20210809T000339_039145_049EAC_rtc', 'S1A_IW_GRDH_1SDV_20210813T120549_20210813T120614_039211_04A0FF_rtc', 'S1A_IW_GRDH_1SDV_20210818T121356_20210818T121421_039284_04A383_rtc', 'S1A_IW_GRDH_1SDV_20210821T000315_20210821T000340_039320_04A4B9_rtc', ... 'S1A_IW_GRDH_1SDV_20220305T120547_20220305T120612_042186_0506F7_rtc', 'S1A_IW_GRDH_1SDV_20220310T121354_20220310T121419_042259_050962_rtc', 'S1A_IW_GRDH_1SDV_20220313T000313_20220313T000338_042295_050AA2_rtc', 'S1A_IW_GRDH_1SDV_20220317T120547_20220317T120612_042361_050CE5_rtc', 'S1A_IW_GRDH_1SDV_20220322T121354_20220322T121419_042434_050F59_rtc', 'S1A_IW_GRDH_1SDV_20220325T000313_20220325T000338_042470_051096_rtc', 'S1A_IW_GRDH_1SDV_20220329T120547_20220329T120612_042536_0512D7_rtc', 'S1A_IW_GRDH_1SDV_20220403T121355_20220403T121420_042609_05154A_rtc', 'S1A_IW_GRDH_1SDV_20220406T000313_20220406T000338_042645_051682_rtc', 'S1A_IW_GRDH_1SDV_20220410T120547_20220410T120612_042711_0518C0_rtc', 'S1A_IW_GRDH_1SDV_20220415T121355_20220415T121420_042784_051B30_rtc', 'S1A_IW_GRDH_1SDV_20220418T000314_20220418T000339_042820_051C63_rtc', 'S1A_IW_GRDH_1SDV_20220422T120548_20220422T120613_042886_051E96_rtc', 'S1A_IW_GRDH_1SDV_20220427T121355_20220427T121420_042959_0520EE_rtc', 'S1A_IW_GRDH_1SDV_20220430T000314_20220430T000339_042995_05221E_rtc', 'S1A_IW_GRDH_1SDV_20220504T120548_20220504T120613_043061_05245F_rtc', 'S1A_IW_GRDH_1SDV_20220509T121356_20220509T121421_043134_0526C4_rtc', 'S1A_IW_GRDH_1SDV_20220516T120549_20220516T120614_043236_0529DD_rtc', 'S1A_IW_GRDH_1SDV_20220521T121357_20220521T121422_043309_052C00_rtc'], dtype='<U66') - band(band)<U2'vh' 'vv'
array(['vh', 'vv'], dtype='<U2')
- x(x)float646.194e+05 6.194e+05 ... 6.281e+05
array([619420., 619430., 619440., ..., 628080., 628090., 628100.])
- y(y)float643.102e+06 3.102e+06 ... 3.09e+06
array([3101660., 3101650., 3101640., ..., 3089810., 3089800., 3089790.])
- sar:looks_azimuth()int641
array(1)
- sat:relative_orbit(time)int64114 12 48 114 12 ... 114 12 114 12
array([114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 114, 12]) - s1:instrument_configuration_ID(time)<U1'6' '6' '6' '6' ... '7' '7' '7' '7'
array(['6', '6', '6', '6', '6', '6', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7'], dtype='<U1') - s1:orbit_source(time)<U8'DOWNLINK' 'DOWNLINK' ... 'RESORB'
array(['DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB'], dtype='<U8') - end_datetime(time)<U32'2021-06-02 12:06:09.940813+00:0...
array(['2021-06-02 12:06:09.940813+00:00', '2021-06-07 12:14:17.557268+00:00', '2021-06-10 00:03:36.369821+00:00', '2021-06-14 12:06:10.872859+00:00', '2021-06-19 12:14:18.365410+00:00', '2021-06-22 00:03:37.093898+00:00', '2021-06-26 12:06:11.482149+00:00', '2021-07-01 12:14:19.071405+00:00', '2021-07-04 00:03:37.847082+00:00', '2021-07-08 12:06:12.112559+00:00', '2021-07-16 00:03:38.592472+00:00', '2021-07-20 12:06:12.927516+00:00', '2021-07-25 12:14:20.313830+00:00', '2021-07-28 00:03:39.166376+00:00', '2021-08-01 12:06:13.649807+00:00', '2021-08-06 12:14:21.197519+00:00', '2021-08-09 00:03:39.953924+00:00', '2021-08-13 12:06:14.213502+00:00', '2021-08-18 12:14:21.725577+00:00', '2021-08-21 00:03:40.559065+00:00', ... '2022-03-01 00:03:38.089393+00:00', '2022-03-05 12:06:12.176227+00:00', '2022-03-10 12:14:19.493699+00:00', '2022-03-13 00:03:38.244245+00:00', '2022-03-17 12:06:12.205265+00:00', '2022-03-22 12:14:19.861261+00:00', '2022-03-25 00:03:38.638828+00:00', '2022-03-29 12:06:12.640119+00:00', '2022-04-03 12:14:20.084951+00:00', '2022-04-06 00:03:38.681256+00:00', '2022-04-10 12:06:12.897487+00:00', '2022-04-15 12:14:20.210307+00:00', '2022-04-18 00:03:39.102936+00:00', '2022-04-22 12:06:13.551041+00:00', '2022-04-27 12:14:20.924939+00:00', '2022-04-30 00:03:39.701264+00:00', '2022-05-04 12:06:13.622093+00:00', '2022-05-09 12:14:21.479341+00:00', '2022-05-16 12:06:14.814536+00:00', '2022-05-21 12:14:22.285304+00:00'], dtype='<U32') - sat:orbit_state(time)<U10'ascending' ... 'ascending'
array(['ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'ascending', 'ascending'], dtype='<U10') - platform()<U11'SENTINEL-1A'
array('SENTINEL-1A', dtype='<U11') - s1:slice_number(time)<U2'6' '6' '12' '6' ... '6' '6' '6'
array(['6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '6', '6'], dtype='<U2') - start_datetime(time)<U32'2021-06-02 12:05:44.941334+00:0...
array(['2021-06-02 12:05:44.941334+00:00', '2021-06-07 12:13:52.559418+00:00', '2021-06-10 00:03:11.371078+00:00', '2021-06-14 12:05:45.874802+00:00', '2021-06-19 12:13:53.365914+00:00', '2021-06-22 00:03:12.095344+00:00', '2021-06-26 12:05:46.482738+00:00', '2021-07-01 12:13:54.073541+00:00', '2021-07-04 00:03:12.848412+00:00', '2021-07-08 12:05:47.114550+00:00', '2021-07-16 00:03:13.593918+00:00', '2021-07-20 12:05:47.929430+00:00', '2021-07-25 12:13:55.316007+00:00', '2021-07-28 00:03:14.167671+00:00', '2021-08-01 12:05:48.651907+00:00', '2021-08-06 12:13:56.199672+00:00', '2021-08-09 00:03:14.955252+00:00', '2021-08-13 12:05:49.214084+00:00', '2021-08-18 12:13:56.727697+00:00', '2021-08-21 00:03:15.560368+00:00', ... '2022-03-01 00:03:13.090580+00:00', '2022-03-05 12:05:47.177919+00:00', '2022-03-10 12:13:54.495512+00:00', '2022-03-13 00:03:13.245549+00:00', '2022-03-17 12:05:47.206984+00:00', '2022-03-22 12:13:54.863098+00:00', '2022-03-25 00:03:13.640217+00:00', '2022-03-29 12:05:47.640405+00:00', '2022-04-03 12:13:55.086898+00:00', '2022-04-06 00:03:13.682743+00:00', '2022-04-10 12:05:47.899396+00:00', '2022-04-15 12:13:55.212427+00:00', '2022-04-18 00:03:14.104239+00:00', '2022-04-22 12:05:48.551577+00:00', '2022-04-27 12:13:55.927061+00:00', '2022-04-30 00:03:14.702542+00:00', '2022-05-04 12:05:48.622638+00:00', '2022-05-09 12:13:56.481455+00:00', '2022-05-16 12:05:49.815129+00:00', '2022-05-21 12:13:57.287447+00:00'], dtype='<U32') - sar:resolution_azimuth()int6422
array(22)
- s1:resolution()<U4'high'
array('high', dtype='<U4') - sar:frequency_band()<U1'C'
array('C', dtype='<U1') - sar:center_frequency()float645.405
array(5.405)
- sar:pixel_spacing_azimuth()int6410
array(10)
- sat:platform_international_designator()<U9'2014-016A'
array('2014-016A', dtype='<U9') - constellation()<U10'Sentinel-1'
array('Sentinel-1', dtype='<U10') - sar:polarizations()object{'VV', 'VH'}
array({'VV', 'VH'}, dtype=object) - proj:epsg()int6432645
array(32645)
- sar:pixel_spacing_range()int6410
array(10)
- sar:looks_range()int645
array(5)
- sar:resolution_range()int6420
array(20)
- sat:absolute_orbit(time)int6438161 38234 38270 ... 43236 43309
array([38161, 38234, 38270, 38336, 38409, 38445, 38511, 38584, 38620, 38686, 38795, 38861, 38934, 38970, 39036, 39109, 39145, 39211, 39284, 39320, 39386, 39495, 39561, 39634, 39670, 39736, 39809, 39845, 39911, 40020, 40086, 40159, 40195, 40261, 40334, 40370, 40436, 40509, 40545, 40611, 40684, 40720, 40786, 40859, 40895, 40961, 41034, 41070, 41136, 41209, 41245, 41311, 41384, 41420, 41486, 41559, 41595, 41661, 41734, 41909, 41945, 42011, 42084, 42120, 42186, 42259, 42295, 42361, 42434, 42470, 42536, 42609, 42645, 42711, 42784, 42820, 42886, 42959, 42995, 43061, 43134, 43236, 43309]) - sar:looks_equivalent_number()float644.4
array(4.4)
- sar:observation_direction()<U5'right'
array('right', dtype='<U5') - sar:product_type()<U3'GRD'
array('GRD', dtype='<U3') - s1:product_timeliness()<U8'Fast-24h'
array('Fast-24h', dtype='<U8') - sar:instrument_mode()<U2'IW'
array('IW', dtype='<U2') - s1:datatake_id(time)<U6'295165' '295704' ... '338944'
array(['295165', '295704', '295966', '296495', '297036', '297306', '297836', '298375', '298649', '299181', '299995', '300524', '301071', '301342', '301855', '302448', '302764', '303359', '304003', '304313', '304892', '305843', '306445', '307077', '307396', '307963', '308582', '308907', '309503', '310456', '311058', '311701', '312022', '312622', '313256', '313577', '314182', '314824', '315135', '315727', '316364', '316686', '317281', '317928', '318241', '318833', '319453', '319760', '320320', '320939', '321239', '321841', '322459', '322765', '323304', '323926', '324244', '324822', '325460', '327023', '327346', '327944', '328566', '328879', '329463', '330082', '330402', '330981', '331609', '331926', '332503', '333130', '333442', '334016', '334640', '334947', '335510', '336110', '336414', '336991', '337604', '338397', '338944'], dtype='<U6') - s1:total_slices(time)<U2'21' '20' '17' ... '20' '21' '20'
array(['21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '21', '20'], dtype='<U2') - s1:processing_level()<U1'1'
array('1', dtype='<U1') - title(band)<U41'VH: vertical transmit, horizont...
array(['VH: vertical transmit, horizontal receive', 'VV: vertical transmit, vertical receive'], dtype='<U41') - description(band)<U173'Terrain-corrected gamma naught ...
array(['Terrain-corrected gamma naught values of signal transmitted with vertical polarization and received with horizontal polarization with radiometric terrain correction applied.', 'Terrain-corrected gamma naught values of signal transmitted with vertical polarization and received with vertical polarization with radiometric terrain correction applied.'], dtype='<U173') - raster:bands()object{'nodata': -32768, 'data_type': ...
array({'nodata': -32768, 'data_type': 'float32', 'spatial_resolution': 10.0}, dtype=object) - epsg()int6432645
array(32645)
- granule_id(time)<U62'S1A_IW_GRDH_1SDV_20210602T12054...
array(['S1A_IW_GRDH_1SDV_20210602T120544_20210602T120609_038161_0480FD', 'S1A_IW_GRDH_1SDV_20210607T121352_20210607T121417_038234_048318', 'S1A_IW_GRDH_1SDV_20210610T000311_20210610T000336_038270_04841E', 'S1A_IW_GRDH_1SDV_20210614T120545_20210614T120610_038336_04862F', 'S1A_IW_GRDH_1SDV_20210619T121353_20210619T121418_038409_04884C', 'S1A_IW_GRDH_1SDV_20210622T000312_20210622T000337_038445_04895A', 'S1A_IW_GRDH_1SDV_20210626T120546_20210626T120611_038511_048B6C', 'S1A_IW_GRDH_1SDV_20210701T121354_20210701T121419_038584_048D87', 'S1A_IW_GRDH_1SDV_20210704T000312_20210704T000337_038620_048E99', 'S1A_IW_GRDH_1SDV_20210708T120547_20210708T120612_038686_0490AD', 'S1A_IW_GRDH_1SDV_20210716T000313_20210716T000338_038795_0493DB', 'S1A_IW_GRDH_1SDV_20210720T120547_20210720T120612_038861_0495EC', 'S1A_IW_GRDH_1SDV_20210725T121355_20210725T121420_038934_04980F', 'S1A_IW_GRDH_1SDV_20210728T000314_20210728T000339_038970_04991E', 'S1A_IW_GRDH_1SDV_20210801T120548_20210801T120613_039036_049B1F', 'S1A_IW_GRDH_1SDV_20210806T121356_20210806T121421_039109_049D70', 'S1A_IW_GRDH_1SDV_20210809T000314_20210809T000339_039145_049EAC', 'S1A_IW_GRDH_1SDV_20210813T120549_20210813T120614_039211_04A0FF', 'S1A_IW_GRDH_1SDV_20210818T121356_20210818T121421_039284_04A383', 'S1A_IW_GRDH_1SDV_20210821T000315_20210821T000340_039320_04A4B9', ... 'S1A_IW_GRDH_1SDV_20220305T120547_20220305T120612_042186_0506F7', 'S1A_IW_GRDH_1SDV_20220310T121354_20220310T121419_042259_050962', 'S1A_IW_GRDH_1SDV_20220313T000313_20220313T000338_042295_050AA2', 'S1A_IW_GRDH_1SDV_20220317T120547_20220317T120612_042361_050CE5', 'S1A_IW_GRDH_1SDV_20220322T121354_20220322T121419_042434_050F59', 'S1A_IW_GRDH_1SDV_20220325T000313_20220325T000338_042470_051096', 'S1A_IW_GRDH_1SDV_20220329T120547_20220329T120612_042536_0512D7', 'S1A_IW_GRDH_1SDV_20220403T121355_20220403T121420_042609_05154A', 'S1A_IW_GRDH_1SDV_20220406T000313_20220406T000338_042645_051682', 'S1A_IW_GRDH_1SDV_20220410T120547_20220410T120612_042711_0518C0', 'S1A_IW_GRDH_1SDV_20220415T121355_20220415T121420_042784_051B30', 'S1A_IW_GRDH_1SDV_20220418T000314_20220418T000339_042820_051C63', 'S1A_IW_GRDH_1SDV_20220422T120548_20220422T120613_042886_051E96', 'S1A_IW_GRDH_1SDV_20220427T121355_20220427T121420_042959_0520EE', 'S1A_IW_GRDH_1SDV_20220430T000314_20220430T000339_042995_05221E', 'S1A_IW_GRDH_1SDV_20220504T120548_20220504T120613_043061_05245F', 'S1A_IW_GRDH_1SDV_20220509T121356_20220509T121421_043134_0526C4', 'S1A_IW_GRDH_1SDV_20220516T120549_20220516T120614_043236_0529DD', 'S1A_IW_GRDH_1SDV_20220521T121357_20220521T121422_043309_052C00'], dtype='<U62') - data_take_id(time)<U6'0480FD' '048318' ... '052C00'
array(['0480FD', '048318', '04841E', '04862F', '04884C', '04895A', '048B6C', '048D87', '048E99', '0490AD', '0493DB', '0495EC', '04980F', '04991E', '049B1F', '049D70', '049EAC', '04A0FF', '04A383', '04A4B9', '04A6FC', '04AAB3', '04AD0D', '04AF85', '04B0C4', '04B2FB', '04B566', '04B6AB', '04B8FF', '04BCB8', '04BF12', '04C195', '04C2D6', '04C52E', '04C7A8', '04C8E9', '04CB46', '04CDC8', '04CEFF', '04D14F', '04D3CC', '04D50E', '04D761', '04D9E8', '04DB21', '04DD71', '04DFDD', '04E110', '04E340', '04E5AB', '04E6D7', '04E931', '04EB9B', '04ECCD', '04EEE8', '04F156', '04F294', '04F4D6', '04F754', '04FD6F', '04FEB2', '050108', '050376', '0504AF', '0506F7', '050962', '050AA2', '050CE5', '050F59', '051096', '0512D7', '05154A', '051682', '0518C0', '051B30', '051C63', '051E96', '0520EE', '05221E', '05245F', '0526C4', '0529DD', '052C00'], dtype='<U6')
- spec :
- RasterSpec(epsg=32645, bounds=(619420.0, 3089780.0, 628110.0, 3101660.0), resolutions_xy=(10.0, 10.0))
- crs :
- epsg:32645
- transform :
- | 10.00, 0.00, 619420.00| | 0.00,-10.00, 3101660.00| | 0.00, 0.00, 1.00|
- resolution :
- 10.0
asf_subset
<xarray.Dataset>
Dimensions: (acq_date: 83, y: 396, x: 290)
Coordinates: (12/20)
* acq_date (acq_date) datetime64[ns] 2021-06-02 .....
granule_id (acq_date) <U67 'S1A_IW_SLC__1SDV_20210...
* x (x) float64 6.194e+05 ... 6.281e+05
* y (y) float64 3.102e+06 ... 3.09e+06
spatial_ref int64 0
sensor (acq_date) <U3 'S1A' 'S1A' ... 'S1A' 'S1A'
... ...
filtered (acq_date) <U1 'n' 'n' 'n' ... 'n' 'n' 'n'
area (acq_date) <U1 'e' 'e' 'e' ... 'e' 'e' 'e'
product_id (acq_date) <U4 '1424' 'ABA0' ... 'BD36'
acq_hour (acq_date) int64 12 12 0 12 0 ... 0 0 0 0
orbital_dir (acq_date) <U4 'desc' 'desc' ... 'asc'
data_take_id (acq_date) <U6 '0480FD' ... '052C00'
Data variables:
vv (acq_date, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
vh (acq_date, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
ls (acq_date, y, x) float64 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>- acq_date: 83
- y: 396
- x: 290
- acq_date(acq_date)datetime64[ns]2021-06-02 ... 2022-05-21
array(['2021-06-02T00:00:00.000000000', '2021-06-07T00:00:00.000000000', '2021-06-10T00:00:00.000000000', '2021-06-14T00:00:00.000000000', '2021-06-19T00:00:00.000000000', '2021-06-22T00:00:00.000000000', '2021-06-26T00:00:00.000000000', '2021-07-01T00:00:00.000000000', '2021-07-04T00:00:00.000000000', '2021-07-08T00:00:00.000000000', '2021-07-16T00:00:00.000000000', '2021-07-20T00:00:00.000000000', '2021-07-25T00:00:00.000000000', '2021-07-28T00:00:00.000000000', '2021-08-01T00:00:00.000000000', '2021-08-06T00:00:00.000000000', '2021-08-09T00:00:00.000000000', '2021-08-13T00:00:00.000000000', '2021-08-18T00:00:00.000000000', '2021-08-21T00:00:00.000000000', '2021-08-25T00:00:00.000000000', '2021-09-02T00:00:00.000000000', '2021-09-06T00:00:00.000000000', '2021-09-11T00:00:00.000000000', '2021-09-14T00:00:00.000000000', '2021-09-18T00:00:00.000000000', '2021-09-23T00:00:00.000000000', '2021-09-26T00:00:00.000000000', '2021-09-30T00:00:00.000000000', '2021-10-08T00:00:00.000000000', '2021-10-12T00:00:00.000000000', '2021-10-17T00:00:00.000000000', '2021-10-20T00:00:00.000000000', '2021-10-24T00:00:00.000000000', '2021-10-29T00:00:00.000000000', '2021-11-01T00:00:00.000000000', '2021-11-05T00:00:00.000000000', '2021-11-10T00:00:00.000000000', '2021-11-13T00:00:00.000000000', '2021-11-17T00:00:00.000000000', '2021-11-22T00:00:00.000000000', '2021-11-25T00:00:00.000000000', '2021-11-29T00:00:00.000000000', '2021-12-04T00:00:00.000000000', '2021-12-07T00:00:00.000000000', '2021-12-11T00:00:00.000000000', '2021-12-16T00:00:00.000000000', '2021-12-19T00:00:00.000000000', '2021-12-23T00:00:00.000000000', '2021-12-28T00:00:00.000000000', '2021-12-31T00:00:00.000000000', '2022-01-04T00:00:00.000000000', '2022-01-09T00:00:00.000000000', '2022-01-12T00:00:00.000000000', '2022-01-16T00:00:00.000000000', '2022-01-21T00:00:00.000000000', '2022-01-24T00:00:00.000000000', '2022-01-28T00:00:00.000000000', '2022-02-02T00:00:00.000000000', '2022-02-14T00:00:00.000000000', '2022-02-17T00:00:00.000000000', '2022-02-21T00:00:00.000000000', '2022-02-26T00:00:00.000000000', '2022-03-01T00:00:00.000000000', '2022-03-05T00:00:00.000000000', '2022-03-10T00:00:00.000000000', '2022-03-13T00:00:00.000000000', '2022-03-17T00:00:00.000000000', '2022-03-22T00:00:00.000000000', '2022-03-25T00:00:00.000000000', '2022-03-29T00:00:00.000000000', '2022-04-03T00:00:00.000000000', '2022-04-06T00:00:00.000000000', '2022-04-10T00:00:00.000000000', '2022-04-15T00:00:00.000000000', '2022-04-18T00:00:00.000000000', '2022-04-22T00:00:00.000000000', '2022-04-27T00:00:00.000000000', '2022-04-30T00:00:00.000000000', '2022-05-04T00:00:00.000000000', '2022-05-09T00:00:00.000000000', '2022-05-16T00:00:00.000000000', '2022-05-21T00:00:00.000000000'], dtype='datetime64[ns]') - granule_id(acq_date)<U67'S1A_IW_SLC__1SDV_20210602T12054...
- description :
- source granule ID for ASF-processed S1 RTC imagery, extracted from README files for each scene
array(['S1A_IW_SLC__1SDV_20210602T120543_20210602T120610_038161_0480FD_893F', 'S1A_IW_SLC__1SDV_20210607T121351_20210607T121418_038234_048318_9B64', 'S1A_IW_SLC__1SDV_20210610T000310_20210610T000337_038270_04841E_601A', 'S1A_IW_SLC__1SDV_20210614T120544_20210614T120611_038336_04862F_337B', 'S1A_IW_SLC__1SDV_20210619T121352_20210619T121419_038409_04884C_1DBA', 'S1A_IW_SLC__1SDV_20210622T000310_20210622T000337_038445_04895A_842E', 'S1A_IW_SLC__1SDV_20210626T120545_20210626T120612_038511_048B6C_9585', 'S1A_IW_SLC__1SDV_20210701T121352_20210701T121419_038584_048D87_F95C', 'S1A_IW_SLC__1SDV_20210704T000311_20210704T000338_038620_048E99_1BAA', 'S1A_IW_SLC__1SDV_20210708T120545_20210708T120612_038686_0490AD_DD9E', 'S1A_IW_SLC__1SDV_20210716T000312_20210716T000339_038795_0493DB_53CD', 'S1A_IW_SLC__1SDV_20210720T120546_20210720T120613_038861_0495EC_C9D4', 'S1A_IW_SLC__1SDV_20210725T121354_20210725T121421_038934_04980F_1651', 'S1A_IW_SLC__1SDV_20210728T000312_20210728T000339_038970_04991E_C669', 'S1A_IW_SLC__1SDV_20210801T120547_20210801T120614_039036_049B1F_9997', 'S1A_IW_SLC__1SDV_20210806T121354_20210806T121421_039109_049D70_B0A0', 'S1A_IW_SLC__1SDV_20210809T000313_20210809T000340_039145_049EAC_AA8E', 'S1A_IW_SLC__1SDV_20210813T120548_20210813T120614_039211_04A0FF_405F', 'S1A_IW_SLC__1SDV_20210818T121355_20210818T121422_039284_04A383_54D5', 'S1A_IW_SLC__1SDV_20210821T000314_20210821T000341_039320_04A4B9_0878', ... 'S1A_IW_SLC__1SDV_20220305T120545_20220305T120612_042186_0506F7_1601', 'S1A_IW_SLC__1SDV_20220310T121353_20220310T121420_042259_050962_068B', 'S1A_IW_SLC__1SDV_20220313T000311_20220313T000338_042295_050AA2_2909', 'S1A_IW_SLC__1SDV_20220317T120545_20220317T120612_042361_050CE5_4CCF', 'S1A_IW_SLC__1SDV_20220322T121353_20220322T121420_042434_050F59_A1B6', 'S1A_IW_SLC__1SDV_20220325T000312_20220325T000339_042470_051096_BA6F', 'S1A_IW_SLC__1SDV_20220329T120546_20220329T120613_042536_0512D7_425A', 'S1A_IW_SLC__1SDV_20220403T121353_20220403T121420_042609_05154A_3FE7', 'S1A_IW_SLC__1SDV_20220406T000312_20220406T000339_042645_051682_4B40', 'S1A_IW_SLC__1SDV_20220410T120546_20220410T120613_042711_0518C0_3349', 'S1A_IW_SLC__1SDV_20220415T121353_20220415T121420_042784_051B30_EB87', 'S1A_IW_SLC__1SDV_20220418T000312_20220418T000339_042820_051C63_CAB2', 'S1A_IW_SLC__1SDV_20220422T120547_20220422T120614_042886_051E96_41EE', 'S1A_IW_SLC__1SDV_20220427T121354_20220427T121421_042959_0520EE_A42D', 'S1A_IW_SLC__1SDV_20220430T000313_20220430T000340_042995_05221E_46A1', 'S1A_IW_SLC__1SDV_20220504T120547_20220504T120614_043061_05245F_6F62', 'S1A_IW_SLC__1SDV_20220509T121355_20220509T121422_043134_0526C4_092E', 'S1A_IW_SLC__1SDV_20220516T120548_20220516T120615_043236_0529DD_42D2', 'S1A_IW_SLC__1SDV_20220521T121356_20220521T121423_043309_052C00_FE88'], dtype='<U67') - x(x)float646.194e+05 6.195e+05 ... 6.281e+05
- axis :
- X
- long_name :
- x coordinate of projection
- standard_name :
- projection_x_coordinate
- units :
- metre
array([619425., 619455., 619485., ..., 628035., 628065., 628095.])
- y(y)float643.102e+06 3.102e+06 ... 3.09e+06
- axis :
- Y
- long_name :
- y coordinate of projection
- standard_name :
- projection_y_coordinate
- units :
- metre
array([3101655., 3101625., 3101595., ..., 3089865., 3089835., 3089805.])
- spatial_ref()int640
- crs_wkt :
- PROJCS["WGS 84 / UTM zone 45N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",87],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32645"]]
- semi_major_axis :
- 6378137.0
- semi_minor_axis :
- 6356752.314245179
- inverse_flattening :
- 298.257223563
- reference_ellipsoid_name :
- WGS 84
- longitude_of_prime_meridian :
- 0.0
- prime_meridian_name :
- Greenwich
- geographic_crs_name :
- WGS 84
- horizontal_datum_name :
- World Geodetic System 1984
- projected_crs_name :
- WGS 84 / UTM zone 45N
- grid_mapping_name :
- transverse_mercator
- latitude_of_projection_origin :
- 0.0
- longitude_of_central_meridian :
- 87.0
- false_easting :
- 500000.0
- false_northing :
- 0.0
- scale_factor_at_central_meridian :
- 0.9996
- spatial_ref :
- PROJCS["WGS 84 / UTM zone 45N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",87],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32645"]]
- GeoTransform :
- 619410.0 30.0 0.0 3101670.0 0.0 -30.0
array(0)
- sensor(acq_date)<U3'S1A' 'S1A' 'S1A' ... 'S1A' 'S1A'
array(['S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A'], dtype='<U3') - beam_mode(acq_date)<U2'IW' 'IW' 'IW' ... 'IW' 'IW' 'IW'
array(['IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW'], dtype='<U2') - acquisition_time(acq_date)datetime64[ns]2022-10-21T12:14:14 ... 2022-10-...
array(['2022-10-21T12:14:14.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:14:20.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T12:05:43.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:05:43.000000000', '2022-10-21T00:03:14.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:52.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T12:13:50.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T12:14:17.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T00:03:10.000000000', '2022-10-21T12:05:45.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:05:50.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:52.000000000', '2022-10-21T00:03:08.000000000', ... '2022-10-21T12:05:42.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:07.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:50.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T00:03:14.000000000', '2022-10-21T12:05:44.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:13:49.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:45.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T00:03:16.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T12:13:51.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T00:03:09.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T00:03:12.000000000'], dtype='datetime64[ns]') - polarisation_type(acq_date)<U2'DV' 'DV' 'DV' ... 'DV' 'DV' 'DV'
array(['DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV'], dtype='<U2') - orbit_type(acq_date)<U1'P' 'P' 'P' 'P' ... 'P' 'P' 'P' 'P'
array(['P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P'], dtype='<U1') - terrain_correction_pixel_spacing(acq_date)<U5'RTC30' 'RTC30' ... 'RTC30' 'RTC30'
array(['RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30'], dtype='<U5') - output_format(acq_date)<U1'g' 'g' 'g' 'g' ... 'g' 'g' 'g' 'g'
array(['g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g'], dtype='<U1') - output_type(acq_date)<U1'p' 'p' 'p' 'p' ... 'p' 'p' 'p' 'p'
array(['p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p'], dtype='<U1') - masked(acq_date)<U1'u' 'u' 'u' 'u' ... 'u' 'u' 'u' 'u'
array(['u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u'], dtype='<U1') - filtered(acq_date)<U1'n' 'n' 'n' 'n' ... 'n' 'n' 'n' 'n'
array(['n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n'], dtype='<U1') - area(acq_date)<U1'e' 'e' 'e' 'e' ... 'e' 'e' 'e' 'e'
array(['e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e'], dtype='<U1') - product_id(acq_date)<U4'1424' 'ABA0' ... '5BAD' 'BD36'
array(['1424', 'ABA0', 'E61E', 'E5B6', '5569', '71C9', 'B942', 'DCE1', '6581', '1DFD', '01B2', '9849', 'CFFC', '57F2', 'D4E6', 'B7B0', '3770', '52BB', '0579', 'AA68', '24B8', '5FF4', '5F4E', '199D', '0031', 'D082', 'E74B', 'F7ED', '65E0', '528F', '51E7', 'F5AA', 'AC38', '772E', 'F0A6', '066E', '5844', '8CD8', '6340', 'D35A', '056C', '6E6F', '0CAD', '6270', '8A4F', 'BEAB', '54B1', '8F7B', 'F9A9', 'AA59', '57E1', '81EB', '250E', '3C39', '3A2A', '9331', 'C14C', 'F1DE', '106F', 'FD00', '5D96', '971C', 'F573', 'C890', '68EA', '83D7', 'E113', 'D846', 'F4B3', 'BB26', '485C', '6683', '6153', '832B', '7418', '378D', '33F5', '1139', 'B661', '9A2F', '6943', '5BAD', 'BD36'], dtype='<U4') - acq_hour(acq_date)int6412 12 0 12 0 12 0 ... 12 12 0 0 0 0
array([12, 12, 0, 12, 0, 12, 0, 12, 0, 12, 0, 12, 12, 12, 0, 0, 12, 0, 12, 12, 12, 12, 0, 0, 12, 0, 12, 12, 12, 12, 12, 12, 0, 12, 12, 12, 12, 0, 12, 0, 12, 12, 12, 12, 12, 12, 0, 12, 12, 0, 12, 12, 12, 12, 12, 0, 12, 12, 0, 12, 12, 12, 12, 12, 0, 12, 12, 0, 0, 0, 12, 12, 12, 12, 12, 12, 12, 12, 12, 0, 0, 0, 0]) - orbital_dir(acq_date)<U4'desc' 'desc' 'asc' ... 'asc' 'asc'
array(['desc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'asc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'asc', 'asc'], dtype='<U4') - data_take_id(acq_date)<U6'0480FD' '048318' ... '052C00'
- ID of data take from SAR acquisition :
- ID data take
array(['0480FD', '048318', '04841E', '04862F', '04884C', '04895A', '048B6C', '048D87', '048E99', '0490AD', '0493DB', '0495EC', '04980F', '04991E', '049B1F', '049D70', '049EAC', '04A0FF', '04A383', '04A4B9', '04A6FC', '04AAB3', '04AD0D', '04AF85', '04B0C4', '04B2FB', '04B566', '04B6AB', '04B8FF', '04BCB8', '04BF12', '04C195', '04C2D6', '04C52E', '04C7A8', '04C8E9', '04CB46', '04CDC8', '04CEFF', '04D14F', '04D3CC', '04D50E', '04D761', '04D9E8', '04DB21', '04DD71', '04DFDD', '04E110', '04E340', '04E5AB', '04E6D7', '04E931', '04EB9B', '04ECCD', '04EEE8', '04F156', '04F294', '04F4D6', '04F754', '04FD6F', '04FEB2', '050108', '050376', '0504AF', '0506F7', '050962', '050AA2', '050CE5', '050F59', '051096', '0512D7', '05154A', '051682', '0518C0', '051B30', '051C63', '051E96', '0520EE', '05221E', '05245F', '0526C4', '0529DD', '052C00'], dtype='<U6')
- vv(acq_date, y, x)float32dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
- scale_factor :
- 1.0
- add_offset :
- 0.0
- _FillValue :
- 0.0
Array Chunk Bytes 36.36 MiB 448.59 kiB Shape (83, 396, 290) (1, 396, 290) Count 19 Graph Layers 83 Chunks Type float32 numpy.ndarray - vh(acq_date, y, x)float32dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
- scale_factor :
- 1.0
- add_offset :
- 0.0
- _FillValue :
- 0.0
Array Chunk Bytes 36.36 MiB 448.59 kiB Shape (83, 396, 290) (1, 396, 290) Count 30 Graph Layers 83 Chunks Type float32 numpy.ndarray - ls(acq_date, y, x)float64dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
- scale_factor :
- 1.0
- add_offset :
- 0.0
- _FillValue :
- 0
Array Chunk Bytes 72.72 MiB 897.19 kiB Shape (83, 396, 290) (1, 396, 290) Count 34 Graph Layers 83 Chunks Type float64 numpy.ndarray
Visual comparison#
Let’s perform some preliminary visual comparisons of the datasets:
Individual time steps#
Use asf_bc_sidebyside() to compare snapshot views of backscatter at individual time steps:
asf_pc_sidebyside(asf_subset, pc_subset, 2)
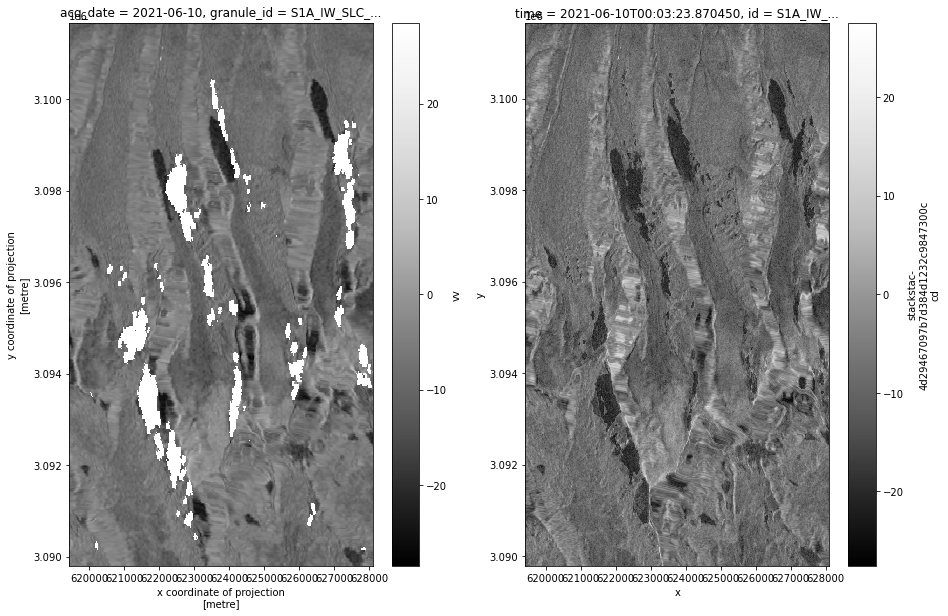
asf_pc_sidebyside(asf_subset, pc_subset, 4)
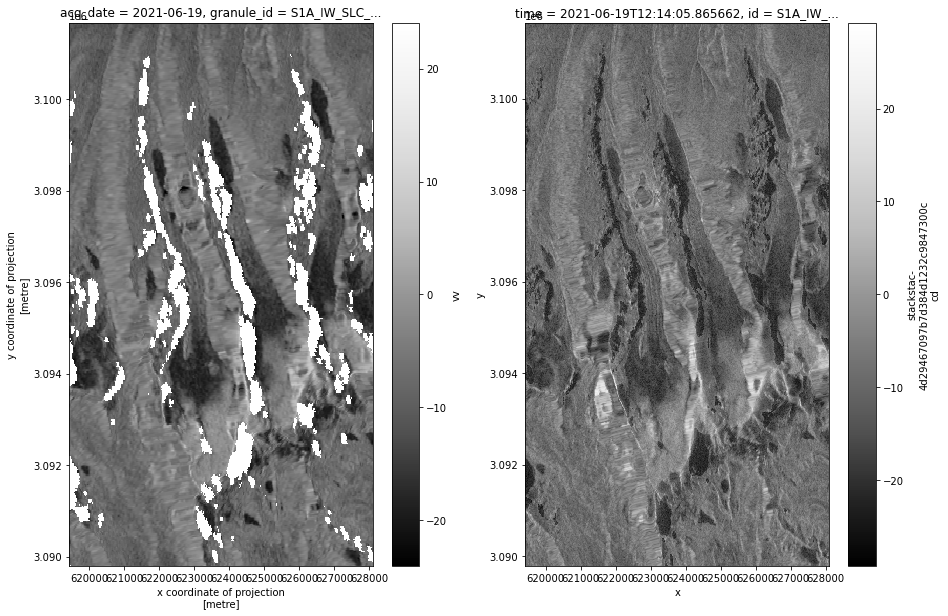
asf_pc_sidebyside(asf_subset, pc_subset, 7)
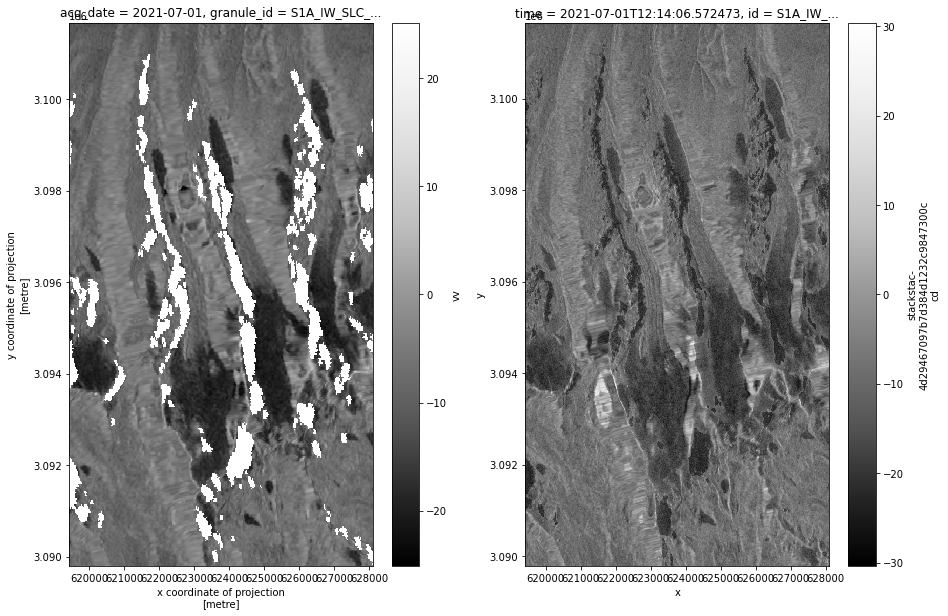
asf_pc_sidebyside(asf_subset, pc_subset, 12)
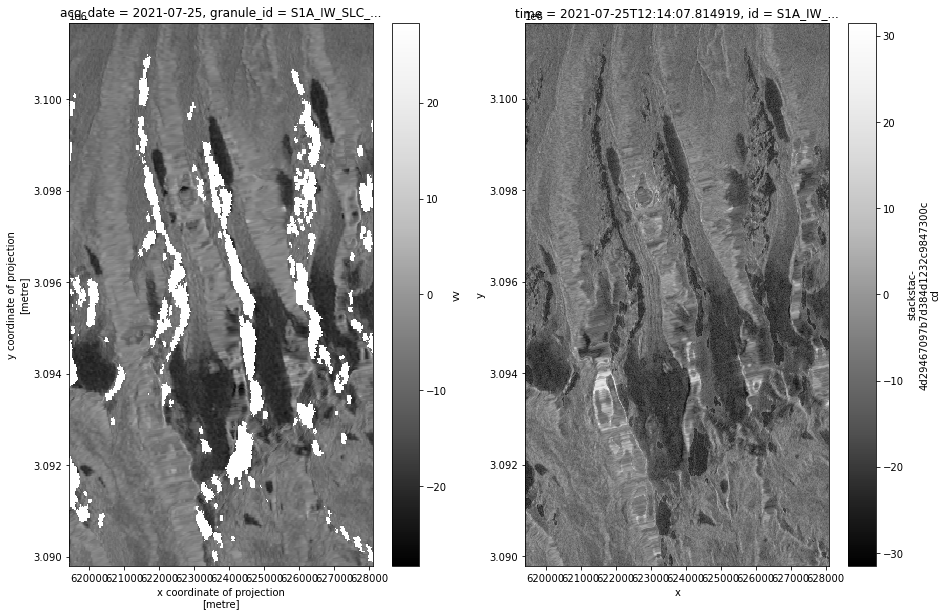
Average backscatter over time#
Now we will look at the backscatter mean along the time dimension
fig, ax = plt.subplots(figsize=(12,8))
power_to_db(asf_subset.vv.mean(dim=['x','y'])).plot(ax=ax, color='blue', linestyle='None', marker='o', markersize=5, label = 'ASF')
power_to_db(pc_subset.sel(band='vv').mean(dim=['x','y'])).plot(ax=ax, color='red', linestyle='None', marker='o', markersize=5, label='PC');
[<matplotlib.lines.Line2D>]
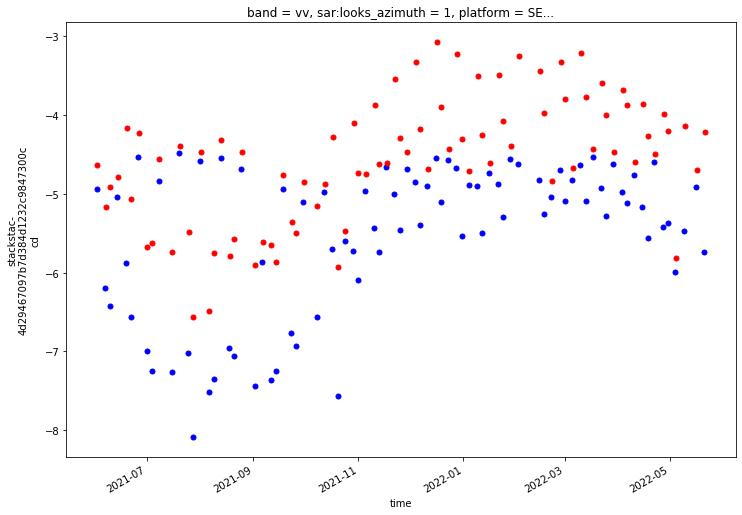
Handling spatial resolution differences#
The PC dataset has a higher spatial resolution than the ASF dataset, which you can see in the sizes of the x and y dimensions of the dataset. The ASF dataset masks out more pixels due to shadow than the PC dataset. To explore whether or not this is the cause of the observed backscatter offset between the two, we need to mask out the pixels in the PC dataset that are masked in the ASF dataset.
In order to do this, first we must downsample the PC dataset to match the spatial resolution of the ASF dataset. There are multiple xarray methods that could be used but here we will use the xr.interp_like() method because we would like the PC x and y dimensions to match the ASF dimensiosn exactly.
pc_downsample = pc_subset.interp_like(asf_subset)
pc_downsample
<xarray.DataArray 'stackstac-4d29467097b7d384d1232c9847300ccd' (time: 83,
band: 2,
y: 396, x: 290)>
dask.array<transpose, shape=(83, 2, 396, 290), dtype=float64, chunksize=(1, 1, 396, 290), chunktype=numpy.ndarray>
Coordinates: (12/41)
* time (time) datetime64[ns] 2021-06-02T1...
id (time) <U66 'S1A_IW_GRDH_1SDV_2021...
* band (band) <U2 'vh' 'vv'
sar:looks_azimuth int64 1
sat:relative_orbit (time) int64 114 12 48 ... 12 114 12
s1:instrument_configuration_ID (time) <U1 '6' '6' '6' ... '7' '7'
... ...
raster:bands object {'nodata': -32768, 'data_ty...
epsg int64 32645
granule_id (time) <U62 'S1A_IW_GRDH_1SDV_2021...
data_take_id (time) <U6 '0480FD' ... '052C00'
* x (x) float64 6.194e+05 ... 6.281e+05
* y (y) float64 3.102e+06 ... 3.09e+06
Attributes:
spec: RasterSpec(epsg=32645, bounds=(619420.0, 3089780.0, 628110.0...
crs: epsg:32645
transform: | 10.00, 0.00, 619420.00|\n| 0.00,-10.00, 3101660.00|\n| 0.0...
resolution: 10.0- time: 83
- band: 2
- y: 396
- x: 290
- dask.array<chunksize=(1, 1, 396, 290), meta=np.ndarray>
Array Chunk Bytes 145.44 MiB 897.19 kiB Shape (83, 2, 396, 290) (1, 1, 396, 290) Count 18 Graph Layers 166 Chunks Type float64 numpy.ndarray - time(time)datetime64[ns]2021-06-02T12:05:57.441074 ... 2...
array(['2021-06-02T12:05:57.441074000', '2021-06-07T12:14:05.058343000', '2021-06-10T00:03:23.870450000', '2021-06-14T12:05:58.373830000', '2021-06-19T12:14:05.865662000', '2021-06-22T00:03:24.594621000', '2021-06-26T12:05:58.982444000', '2021-07-01T12:14:06.572473000', '2021-07-04T00:03:25.347747000', '2021-07-08T12:05:59.613554000', '2021-07-16T00:03:26.093195000', '2021-07-20T12:06:00.428473000', '2021-07-25T12:14:07.814919000', '2021-07-28T00:03:26.667023000', '2021-08-01T12:06:01.150857000', '2021-08-06T12:14:08.698596000', '2021-08-09T00:03:27.454588000', '2021-08-13T12:06:01.713793000', '2021-08-18T12:14:09.226637000', '2021-08-21T00:03:28.059716000', '2021-08-25T12:06:02.421490000', '2021-09-02T00:03:28.464707000', '2021-09-06T12:06:02.971474000', '2021-09-11T12:14:10.406593000', '2021-09-14T00:03:29.135941000', '2021-09-18T12:06:03.415938000', '2021-09-23T12:14:10.788780000', '2021-09-26T00:03:29.558327000', '2021-09-30T12:06:03.760613000', '2021-10-08T00:03:29.713275000', '2021-10-12T12:06:03.850150000', '2021-10-17T12:14:11.200359000', '2021-10-20T00:03:29.919736000', '2021-10-24T12:06:03.954256000', '2021-10-29T12:14:11.052769000', '2021-11-01T00:03:29.702246000', '2021-11-05T12:06:03.645002000', '2021-11-10T12:14:10.742983000', '2021-11-13T00:03:29.481869000', '2021-11-17T12:06:03.560526000', '2021-11-22T12:14:10.576899000', '2021-11-25T00:03:29.120003000', '2021-11-29T12:06:03.035677000', '2021-12-04T12:14:10.007158000', '2021-12-07T00:03:28.568419000', '2021-12-11T12:06:02.513966000', '2021-12-16T12:14:09.596874000', '2021-12-19T00:03:28.141449000', '2021-12-23T12:06:01.793721000', '2021-12-28T12:14:08.858303000', '2021-12-31T00:03:27.432317000', '2022-01-04T12:06:01.280484000', '2022-01-09T12:14:08.415805000', '2022-01-12T00:03:27.070182000', '2022-01-16T12:06:00.877035000', '2022-01-21T12:14:07.716165000', '2022-01-24T00:03:26.424832000', '2022-01-28T12:06:00.352862000', '2022-02-02T12:14:07.057390000', '2022-02-14T12:14:07.178103000', '2022-02-17T00:03:25.677942000', '2022-02-21T12:05:59.939806000', '2022-02-26T12:14:07.075742000', '2022-03-01T00:03:25.589986000', '2022-03-05T12:05:59.677073000', '2022-03-10T12:14:06.994606000', '2022-03-13T00:03:25.744897000', '2022-03-17T12:05:59.706124000', '2022-03-22T12:14:07.362180000', '2022-03-25T00:03:26.139523000', '2022-03-29T12:06:00.140262000', '2022-04-03T12:14:07.585924000', '2022-04-06T00:03:26.181999000', '2022-04-10T12:06:00.398442000', '2022-04-15T12:14:07.711367000', '2022-04-18T00:03:26.603587000', '2022-04-22T12:06:01.051309000', '2022-04-27T12:14:08.426000000', '2022-04-30T00:03:27.201903000', '2022-05-04T12:06:01.122366000', '2022-05-09T12:14:08.980398000', '2022-05-16T12:06:02.314833000', '2022-05-21T12:14:09.786375000'], dtype='datetime64[ns]') - id(time)<U66'S1A_IW_GRDH_1SDV_20210602T12054...
array(['S1A_IW_GRDH_1SDV_20210602T120544_20210602T120609_038161_0480FD_rtc', 'S1A_IW_GRDH_1SDV_20210607T121352_20210607T121417_038234_048318_rtc', 'S1A_IW_GRDH_1SDV_20210610T000311_20210610T000336_038270_04841E_rtc', 'S1A_IW_GRDH_1SDV_20210614T120545_20210614T120610_038336_04862F_rtc', 'S1A_IW_GRDH_1SDV_20210619T121353_20210619T121418_038409_04884C_rtc', 'S1A_IW_GRDH_1SDV_20210622T000312_20210622T000337_038445_04895A_rtc', 'S1A_IW_GRDH_1SDV_20210626T120546_20210626T120611_038511_048B6C_rtc', 'S1A_IW_GRDH_1SDV_20210701T121354_20210701T121419_038584_048D87_rtc', 'S1A_IW_GRDH_1SDV_20210704T000312_20210704T000337_038620_048E99_rtc', 'S1A_IW_GRDH_1SDV_20210708T120547_20210708T120612_038686_0490AD_rtc', 'S1A_IW_GRDH_1SDV_20210716T000313_20210716T000338_038795_0493DB_rtc', 'S1A_IW_GRDH_1SDV_20210720T120547_20210720T120612_038861_0495EC_rtc', 'S1A_IW_GRDH_1SDV_20210725T121355_20210725T121420_038934_04980F_rtc', 'S1A_IW_GRDH_1SDV_20210728T000314_20210728T000339_038970_04991E_rtc', 'S1A_IW_GRDH_1SDV_20210801T120548_20210801T120613_039036_049B1F_rtc', 'S1A_IW_GRDH_1SDV_20210806T121356_20210806T121421_039109_049D70_rtc', 'S1A_IW_GRDH_1SDV_20210809T000314_20210809T000339_039145_049EAC_rtc', 'S1A_IW_GRDH_1SDV_20210813T120549_20210813T120614_039211_04A0FF_rtc', 'S1A_IW_GRDH_1SDV_20210818T121356_20210818T121421_039284_04A383_rtc', 'S1A_IW_GRDH_1SDV_20210821T000315_20210821T000340_039320_04A4B9_rtc', ... 'S1A_IW_GRDH_1SDV_20220305T120547_20220305T120612_042186_0506F7_rtc', 'S1A_IW_GRDH_1SDV_20220310T121354_20220310T121419_042259_050962_rtc', 'S1A_IW_GRDH_1SDV_20220313T000313_20220313T000338_042295_050AA2_rtc', 'S1A_IW_GRDH_1SDV_20220317T120547_20220317T120612_042361_050CE5_rtc', 'S1A_IW_GRDH_1SDV_20220322T121354_20220322T121419_042434_050F59_rtc', 'S1A_IW_GRDH_1SDV_20220325T000313_20220325T000338_042470_051096_rtc', 'S1A_IW_GRDH_1SDV_20220329T120547_20220329T120612_042536_0512D7_rtc', 'S1A_IW_GRDH_1SDV_20220403T121355_20220403T121420_042609_05154A_rtc', 'S1A_IW_GRDH_1SDV_20220406T000313_20220406T000338_042645_051682_rtc', 'S1A_IW_GRDH_1SDV_20220410T120547_20220410T120612_042711_0518C0_rtc', 'S1A_IW_GRDH_1SDV_20220415T121355_20220415T121420_042784_051B30_rtc', 'S1A_IW_GRDH_1SDV_20220418T000314_20220418T000339_042820_051C63_rtc', 'S1A_IW_GRDH_1SDV_20220422T120548_20220422T120613_042886_051E96_rtc', 'S1A_IW_GRDH_1SDV_20220427T121355_20220427T121420_042959_0520EE_rtc', 'S1A_IW_GRDH_1SDV_20220430T000314_20220430T000339_042995_05221E_rtc', 'S1A_IW_GRDH_1SDV_20220504T120548_20220504T120613_043061_05245F_rtc', 'S1A_IW_GRDH_1SDV_20220509T121356_20220509T121421_043134_0526C4_rtc', 'S1A_IW_GRDH_1SDV_20220516T120549_20220516T120614_043236_0529DD_rtc', 'S1A_IW_GRDH_1SDV_20220521T121357_20220521T121422_043309_052C00_rtc'], dtype='<U66') - band(band)<U2'vh' 'vv'
array(['vh', 'vv'], dtype='<U2')
- sar:looks_azimuth()int641
array(1)
- sat:relative_orbit(time)int64114 12 48 114 12 ... 114 12 114 12
array([114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 114, 12]) - s1:instrument_configuration_ID(time)<U1'6' '6' '6' '6' ... '7' '7' '7' '7'
array(['6', '6', '6', '6', '6', '6', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7'], dtype='<U1') - s1:orbit_source(time)<U8'DOWNLINK' 'DOWNLINK' ... 'RESORB'
array(['DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB'], dtype='<U8') - end_datetime(time)<U32'2021-06-02 12:06:09.940813+00:0...
array(['2021-06-02 12:06:09.940813+00:00', '2021-06-07 12:14:17.557268+00:00', '2021-06-10 00:03:36.369821+00:00', '2021-06-14 12:06:10.872859+00:00', '2021-06-19 12:14:18.365410+00:00', '2021-06-22 00:03:37.093898+00:00', '2021-06-26 12:06:11.482149+00:00', '2021-07-01 12:14:19.071405+00:00', '2021-07-04 00:03:37.847082+00:00', '2021-07-08 12:06:12.112559+00:00', '2021-07-16 00:03:38.592472+00:00', '2021-07-20 12:06:12.927516+00:00', '2021-07-25 12:14:20.313830+00:00', '2021-07-28 00:03:39.166376+00:00', '2021-08-01 12:06:13.649807+00:00', '2021-08-06 12:14:21.197519+00:00', '2021-08-09 00:03:39.953924+00:00', '2021-08-13 12:06:14.213502+00:00', '2021-08-18 12:14:21.725577+00:00', '2021-08-21 00:03:40.559065+00:00', ... '2022-03-01 00:03:38.089393+00:00', '2022-03-05 12:06:12.176227+00:00', '2022-03-10 12:14:19.493699+00:00', '2022-03-13 00:03:38.244245+00:00', '2022-03-17 12:06:12.205265+00:00', '2022-03-22 12:14:19.861261+00:00', '2022-03-25 00:03:38.638828+00:00', '2022-03-29 12:06:12.640119+00:00', '2022-04-03 12:14:20.084951+00:00', '2022-04-06 00:03:38.681256+00:00', '2022-04-10 12:06:12.897487+00:00', '2022-04-15 12:14:20.210307+00:00', '2022-04-18 00:03:39.102936+00:00', '2022-04-22 12:06:13.551041+00:00', '2022-04-27 12:14:20.924939+00:00', '2022-04-30 00:03:39.701264+00:00', '2022-05-04 12:06:13.622093+00:00', '2022-05-09 12:14:21.479341+00:00', '2022-05-16 12:06:14.814536+00:00', '2022-05-21 12:14:22.285304+00:00'], dtype='<U32') - sat:orbit_state(time)<U10'ascending' ... 'ascending'
array(['ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'ascending', 'ascending'], dtype='<U10') - platform()<U11'SENTINEL-1A'
array('SENTINEL-1A', dtype='<U11') - s1:slice_number(time)<U2'6' '6' '12' '6' ... '6' '6' '6'
array(['6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '6', '6'], dtype='<U2') - start_datetime(time)<U32'2021-06-02 12:05:44.941334+00:0...
array(['2021-06-02 12:05:44.941334+00:00', '2021-06-07 12:13:52.559418+00:00', '2021-06-10 00:03:11.371078+00:00', '2021-06-14 12:05:45.874802+00:00', '2021-06-19 12:13:53.365914+00:00', '2021-06-22 00:03:12.095344+00:00', '2021-06-26 12:05:46.482738+00:00', '2021-07-01 12:13:54.073541+00:00', '2021-07-04 00:03:12.848412+00:00', '2021-07-08 12:05:47.114550+00:00', '2021-07-16 00:03:13.593918+00:00', '2021-07-20 12:05:47.929430+00:00', '2021-07-25 12:13:55.316007+00:00', '2021-07-28 00:03:14.167671+00:00', '2021-08-01 12:05:48.651907+00:00', '2021-08-06 12:13:56.199672+00:00', '2021-08-09 00:03:14.955252+00:00', '2021-08-13 12:05:49.214084+00:00', '2021-08-18 12:13:56.727697+00:00', '2021-08-21 00:03:15.560368+00:00', ... '2022-03-01 00:03:13.090580+00:00', '2022-03-05 12:05:47.177919+00:00', '2022-03-10 12:13:54.495512+00:00', '2022-03-13 00:03:13.245549+00:00', '2022-03-17 12:05:47.206984+00:00', '2022-03-22 12:13:54.863098+00:00', '2022-03-25 00:03:13.640217+00:00', '2022-03-29 12:05:47.640405+00:00', '2022-04-03 12:13:55.086898+00:00', '2022-04-06 00:03:13.682743+00:00', '2022-04-10 12:05:47.899396+00:00', '2022-04-15 12:13:55.212427+00:00', '2022-04-18 00:03:14.104239+00:00', '2022-04-22 12:05:48.551577+00:00', '2022-04-27 12:13:55.927061+00:00', '2022-04-30 00:03:14.702542+00:00', '2022-05-04 12:05:48.622638+00:00', '2022-05-09 12:13:56.481455+00:00', '2022-05-16 12:05:49.815129+00:00', '2022-05-21 12:13:57.287447+00:00'], dtype='<U32') - sar:resolution_azimuth()int6422
array(22)
- s1:resolution()<U4'high'
array('high', dtype='<U4') - sar:frequency_band()<U1'C'
array('C', dtype='<U1') - sar:center_frequency()float645.405
array(5.405)
- sar:pixel_spacing_azimuth()int6410
array(10)
- sat:platform_international_designator()<U9'2014-016A'
array('2014-016A', dtype='<U9') - constellation()<U10'Sentinel-1'
array('Sentinel-1', dtype='<U10') - sar:polarizations()object{'VV', 'VH'}
array({'VV', 'VH'}, dtype=object) - proj:epsg()int6432645
array(32645)
- sar:pixel_spacing_range()int6410
array(10)
- sar:looks_range()int645
array(5)
- sar:resolution_range()int6420
array(20)
- sat:absolute_orbit(time)int6438161 38234 38270 ... 43236 43309
array([38161, 38234, 38270, 38336, 38409, 38445, 38511, 38584, 38620, 38686, 38795, 38861, 38934, 38970, 39036, 39109, 39145, 39211, 39284, 39320, 39386, 39495, 39561, 39634, 39670, 39736, 39809, 39845, 39911, 40020, 40086, 40159, 40195, 40261, 40334, 40370, 40436, 40509, 40545, 40611, 40684, 40720, 40786, 40859, 40895, 40961, 41034, 41070, 41136, 41209, 41245, 41311, 41384, 41420, 41486, 41559, 41595, 41661, 41734, 41909, 41945, 42011, 42084, 42120, 42186, 42259, 42295, 42361, 42434, 42470, 42536, 42609, 42645, 42711, 42784, 42820, 42886, 42959, 42995, 43061, 43134, 43236, 43309]) - sar:looks_equivalent_number()float644.4
array(4.4)
- sar:observation_direction()<U5'right'
array('right', dtype='<U5') - sar:product_type()<U3'GRD'
array('GRD', dtype='<U3') - s1:product_timeliness()<U8'Fast-24h'
array('Fast-24h', dtype='<U8') - sar:instrument_mode()<U2'IW'
array('IW', dtype='<U2') - s1:datatake_id(time)<U6'295165' '295704' ... '338944'
array(['295165', '295704', '295966', '296495', '297036', '297306', '297836', '298375', '298649', '299181', '299995', '300524', '301071', '301342', '301855', '302448', '302764', '303359', '304003', '304313', '304892', '305843', '306445', '307077', '307396', '307963', '308582', '308907', '309503', '310456', '311058', '311701', '312022', '312622', '313256', '313577', '314182', '314824', '315135', '315727', '316364', '316686', '317281', '317928', '318241', '318833', '319453', '319760', '320320', '320939', '321239', '321841', '322459', '322765', '323304', '323926', '324244', '324822', '325460', '327023', '327346', '327944', '328566', '328879', '329463', '330082', '330402', '330981', '331609', '331926', '332503', '333130', '333442', '334016', '334640', '334947', '335510', '336110', '336414', '336991', '337604', '338397', '338944'], dtype='<U6') - s1:total_slices(time)<U2'21' '20' '17' ... '20' '21' '20'
array(['21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '21', '20'], dtype='<U2') - s1:processing_level()<U1'1'
array('1', dtype='<U1') - title(band)<U41'VH: vertical transmit, horizont...
array(['VH: vertical transmit, horizontal receive', 'VV: vertical transmit, vertical receive'], dtype='<U41') - description(band)<U173'Terrain-corrected gamma naught ...
array(['Terrain-corrected gamma naught values of signal transmitted with vertical polarization and received with horizontal polarization with radiometric terrain correction applied.', 'Terrain-corrected gamma naught values of signal transmitted with vertical polarization and received with vertical polarization with radiometric terrain correction applied.'], dtype='<U173') - raster:bands()object{'nodata': -32768, 'data_type': ...
array({'nodata': -32768, 'data_type': 'float32', 'spatial_resolution': 10.0}, dtype=object) - epsg()int6432645
array(32645)
- granule_id(time)<U62'S1A_IW_GRDH_1SDV_20210602T12054...
array(['S1A_IW_GRDH_1SDV_20210602T120544_20210602T120609_038161_0480FD', 'S1A_IW_GRDH_1SDV_20210607T121352_20210607T121417_038234_048318', 'S1A_IW_GRDH_1SDV_20210610T000311_20210610T000336_038270_04841E', 'S1A_IW_GRDH_1SDV_20210614T120545_20210614T120610_038336_04862F', 'S1A_IW_GRDH_1SDV_20210619T121353_20210619T121418_038409_04884C', 'S1A_IW_GRDH_1SDV_20210622T000312_20210622T000337_038445_04895A', 'S1A_IW_GRDH_1SDV_20210626T120546_20210626T120611_038511_048B6C', 'S1A_IW_GRDH_1SDV_20210701T121354_20210701T121419_038584_048D87', 'S1A_IW_GRDH_1SDV_20210704T000312_20210704T000337_038620_048E99', 'S1A_IW_GRDH_1SDV_20210708T120547_20210708T120612_038686_0490AD', 'S1A_IW_GRDH_1SDV_20210716T000313_20210716T000338_038795_0493DB', 'S1A_IW_GRDH_1SDV_20210720T120547_20210720T120612_038861_0495EC', 'S1A_IW_GRDH_1SDV_20210725T121355_20210725T121420_038934_04980F', 'S1A_IW_GRDH_1SDV_20210728T000314_20210728T000339_038970_04991E', 'S1A_IW_GRDH_1SDV_20210801T120548_20210801T120613_039036_049B1F', 'S1A_IW_GRDH_1SDV_20210806T121356_20210806T121421_039109_049D70', 'S1A_IW_GRDH_1SDV_20210809T000314_20210809T000339_039145_049EAC', 'S1A_IW_GRDH_1SDV_20210813T120549_20210813T120614_039211_04A0FF', 'S1A_IW_GRDH_1SDV_20210818T121356_20210818T121421_039284_04A383', 'S1A_IW_GRDH_1SDV_20210821T000315_20210821T000340_039320_04A4B9', ... 'S1A_IW_GRDH_1SDV_20220305T120547_20220305T120612_042186_0506F7', 'S1A_IW_GRDH_1SDV_20220310T121354_20220310T121419_042259_050962', 'S1A_IW_GRDH_1SDV_20220313T000313_20220313T000338_042295_050AA2', 'S1A_IW_GRDH_1SDV_20220317T120547_20220317T120612_042361_050CE5', 'S1A_IW_GRDH_1SDV_20220322T121354_20220322T121419_042434_050F59', 'S1A_IW_GRDH_1SDV_20220325T000313_20220325T000338_042470_051096', 'S1A_IW_GRDH_1SDV_20220329T120547_20220329T120612_042536_0512D7', 'S1A_IW_GRDH_1SDV_20220403T121355_20220403T121420_042609_05154A', 'S1A_IW_GRDH_1SDV_20220406T000313_20220406T000338_042645_051682', 'S1A_IW_GRDH_1SDV_20220410T120547_20220410T120612_042711_0518C0', 'S1A_IW_GRDH_1SDV_20220415T121355_20220415T121420_042784_051B30', 'S1A_IW_GRDH_1SDV_20220418T000314_20220418T000339_042820_051C63', 'S1A_IW_GRDH_1SDV_20220422T120548_20220422T120613_042886_051E96', 'S1A_IW_GRDH_1SDV_20220427T121355_20220427T121420_042959_0520EE', 'S1A_IW_GRDH_1SDV_20220430T000314_20220430T000339_042995_05221E', 'S1A_IW_GRDH_1SDV_20220504T120548_20220504T120613_043061_05245F', 'S1A_IW_GRDH_1SDV_20220509T121356_20220509T121421_043134_0526C4', 'S1A_IW_GRDH_1SDV_20220516T120549_20220516T120614_043236_0529DD', 'S1A_IW_GRDH_1SDV_20220521T121357_20220521T121422_043309_052C00'], dtype='<U62') - data_take_id(time)<U6'0480FD' '048318' ... '052C00'
array(['0480FD', '048318', '04841E', '04862F', '04884C', '04895A', '048B6C', '048D87', '048E99', '0490AD', '0493DB', '0495EC', '04980F', '04991E', '049B1F', '049D70', '049EAC', '04A0FF', '04A383', '04A4B9', '04A6FC', '04AAB3', '04AD0D', '04AF85', '04B0C4', '04B2FB', '04B566', '04B6AB', '04B8FF', '04BCB8', '04BF12', '04C195', '04C2D6', '04C52E', '04C7A8', '04C8E9', '04CB46', '04CDC8', '04CEFF', '04D14F', '04D3CC', '04D50E', '04D761', '04D9E8', '04DB21', '04DD71', '04DFDD', '04E110', '04E340', '04E5AB', '04E6D7', '04E931', '04EB9B', '04ECCD', '04EEE8', '04F156', '04F294', '04F4D6', '04F754', '04FD6F', '04FEB2', '050108', '050376', '0504AF', '0506F7', '050962', '050AA2', '050CE5', '050F59', '051096', '0512D7', '05154A', '051682', '0518C0', '051B30', '051C63', '051E96', '0520EE', '05221E', '05245F', '0526C4', '0529DD', '052C00'], dtype='<U6') - x(x)float646.194e+05 6.195e+05 ... 6.281e+05
- axis :
- X
- long_name :
- x coordinate of projection
- standard_name :
- projection_x_coordinate
- units :
- metre
array([619425., 619455., 619485., ..., 628035., 628065., 628095.])
- y(y)float643.102e+06 3.102e+06 ... 3.09e+06
- axis :
- Y
- long_name :
- y coordinate of projection
- standard_name :
- projection_y_coordinate
- units :
- metre
array([3101655., 3101625., 3101595., ..., 3089865., 3089835., 3089805.])
- spec :
- RasterSpec(epsg=32645, bounds=(619420.0, 3089780.0, 628110.0, 3101660.0), resolutions_xy=(10.0, 10.0))
- crs :
- epsg:32645
- transform :
- | 10.00, 0.00, 619420.00| | 0.00,-10.00, 3101660.00| | 0.00, 0.00, 1.00|
- resolution :
- 10.0
pc_mask = xr.where(asf_subset.notnull(), pc_downsample,np.nan)
/home/emmamarshall/miniconda3/envs/sentinel/lib/python3.10/site-packages/dask/array/core.py:4796: PerformanceWarning: Increasing number of chunks by factor of 83
result = blockwise(
/home/emmamarshall/miniconda3/envs/sentinel/lib/python3.10/site-packages/dask/array/core.py:4796: PerformanceWarning: Increasing number of chunks by factor of 83
result = blockwise(
/home/emmamarshall/miniconda3/envs/sentinel/lib/python3.10/site-packages/dask/array/core.py:4796: PerformanceWarning: Increasing number of chunks by factor of 83
result = blockwise(
pc_mask
<xarray.Dataset>
Dimensions: (acq_date: 83, y: 396, x: 290,
time: 83, band: 2)
Coordinates: (12/55)
* acq_date (acq_date) datetime64[ns] 2021-06-...
* x (x) float64 6.194e+05 ... 6.281e+05
* y (y) float64 3.102e+06 ... 3.09e+06
spatial_ref int64 0
sensor (acq_date) <U3 'S1A' 'S1A' ... 'S1A'
beam_mode (acq_date) <U2 'IW' 'IW' ... 'IW'
... ...
s1:total_slices (time) <U2 '21' '20' ... '21' '20'
s1:processing_level <U1 '1'
title (band) <U41 'VH: vertical transmit...
description (band) <U173 'Terrain-corrected ga...
raster:bands object {'nodata': -32768, 'data_ty...
epsg int64 32645
Data variables:
vv (acq_date, y, x, time, band) float64 dask.array<chunksize=(1, 396, 290, 1, 1), meta=np.ndarray>
vh (acq_date, y, x, time, band) float64 dask.array<chunksize=(1, 396, 290, 1, 1), meta=np.ndarray>
ls (acq_date, y, x, time, band) float64 dask.array<chunksize=(1, 396, 290, 1, 1), meta=np.ndarray>- acq_date: 83
- y: 396
- x: 290
- time: 83
- band: 2
- acq_date(acq_date)datetime64[ns]2021-06-02 ... 2022-05-21
array(['2021-06-02T00:00:00.000000000', '2021-06-07T00:00:00.000000000', '2021-06-10T00:00:00.000000000', '2021-06-14T00:00:00.000000000', '2021-06-19T00:00:00.000000000', '2021-06-22T00:00:00.000000000', '2021-06-26T00:00:00.000000000', '2021-07-01T00:00:00.000000000', '2021-07-04T00:00:00.000000000', '2021-07-08T00:00:00.000000000', '2021-07-16T00:00:00.000000000', '2021-07-20T00:00:00.000000000', '2021-07-25T00:00:00.000000000', '2021-07-28T00:00:00.000000000', '2021-08-01T00:00:00.000000000', '2021-08-06T00:00:00.000000000', '2021-08-09T00:00:00.000000000', '2021-08-13T00:00:00.000000000', '2021-08-18T00:00:00.000000000', '2021-08-21T00:00:00.000000000', '2021-08-25T00:00:00.000000000', '2021-09-02T00:00:00.000000000', '2021-09-06T00:00:00.000000000', '2021-09-11T00:00:00.000000000', '2021-09-14T00:00:00.000000000', '2021-09-18T00:00:00.000000000', '2021-09-23T00:00:00.000000000', '2021-09-26T00:00:00.000000000', '2021-09-30T00:00:00.000000000', '2021-10-08T00:00:00.000000000', '2021-10-12T00:00:00.000000000', '2021-10-17T00:00:00.000000000', '2021-10-20T00:00:00.000000000', '2021-10-24T00:00:00.000000000', '2021-10-29T00:00:00.000000000', '2021-11-01T00:00:00.000000000', '2021-11-05T00:00:00.000000000', '2021-11-10T00:00:00.000000000', '2021-11-13T00:00:00.000000000', '2021-11-17T00:00:00.000000000', '2021-11-22T00:00:00.000000000', '2021-11-25T00:00:00.000000000', '2021-11-29T00:00:00.000000000', '2021-12-04T00:00:00.000000000', '2021-12-07T00:00:00.000000000', '2021-12-11T00:00:00.000000000', '2021-12-16T00:00:00.000000000', '2021-12-19T00:00:00.000000000', '2021-12-23T00:00:00.000000000', '2021-12-28T00:00:00.000000000', '2021-12-31T00:00:00.000000000', '2022-01-04T00:00:00.000000000', '2022-01-09T00:00:00.000000000', '2022-01-12T00:00:00.000000000', '2022-01-16T00:00:00.000000000', '2022-01-21T00:00:00.000000000', '2022-01-24T00:00:00.000000000', '2022-01-28T00:00:00.000000000', '2022-02-02T00:00:00.000000000', '2022-02-14T00:00:00.000000000', '2022-02-17T00:00:00.000000000', '2022-02-21T00:00:00.000000000', '2022-02-26T00:00:00.000000000', '2022-03-01T00:00:00.000000000', '2022-03-05T00:00:00.000000000', '2022-03-10T00:00:00.000000000', '2022-03-13T00:00:00.000000000', '2022-03-17T00:00:00.000000000', '2022-03-22T00:00:00.000000000', '2022-03-25T00:00:00.000000000', '2022-03-29T00:00:00.000000000', '2022-04-03T00:00:00.000000000', '2022-04-06T00:00:00.000000000', '2022-04-10T00:00:00.000000000', '2022-04-15T00:00:00.000000000', '2022-04-18T00:00:00.000000000', '2022-04-22T00:00:00.000000000', '2022-04-27T00:00:00.000000000', '2022-04-30T00:00:00.000000000', '2022-05-04T00:00:00.000000000', '2022-05-09T00:00:00.000000000', '2022-05-16T00:00:00.000000000', '2022-05-21T00:00:00.000000000'], dtype='datetime64[ns]') - x(x)float646.194e+05 6.195e+05 ... 6.281e+05
array([619425., 619455., 619485., ..., 628035., 628065., 628095.])
- y(y)float643.102e+06 3.102e+06 ... 3.09e+06
array([3101655., 3101625., 3101595., ..., 3089865., 3089835., 3089805.])
- spatial_ref()int640
array(0)
- sensor(acq_date)<U3'S1A' 'S1A' 'S1A' ... 'S1A' 'S1A'
array(['S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A'], dtype='<U3') - beam_mode(acq_date)<U2'IW' 'IW' 'IW' ... 'IW' 'IW' 'IW'
array(['IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW'], dtype='<U2') - acquisition_time(acq_date)datetime64[ns]2022-10-21T12:14:14 ... 2022-10-...
array(['2022-10-21T12:14:14.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:14:20.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T12:05:43.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:05:43.000000000', '2022-10-21T00:03:14.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:52.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T12:13:50.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T12:14:17.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T00:03:10.000000000', '2022-10-21T12:05:45.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:05:50.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:52.000000000', '2022-10-21T00:03:08.000000000', ... '2022-10-21T12:05:42.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:07.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:50.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T00:03:14.000000000', '2022-10-21T12:05:44.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:13:49.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:45.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T00:03:16.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T12:13:51.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T00:03:09.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T00:03:12.000000000'], dtype='datetime64[ns]') - polarisation_type(acq_date)<U2'DV' 'DV' 'DV' ... 'DV' 'DV' 'DV'
array(['DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV'], dtype='<U2') - orbit_type(acq_date)<U1'P' 'P' 'P' 'P' ... 'P' 'P' 'P' 'P'
array(['P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P'], dtype='<U1') - terrain_correction_pixel_spacing(acq_date)<U5'RTC30' 'RTC30' ... 'RTC30' 'RTC30'
array(['RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30'], dtype='<U5') - output_format(acq_date)<U1'g' 'g' 'g' 'g' ... 'g' 'g' 'g' 'g'
array(['g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g'], dtype='<U1') - output_type(acq_date)<U1'p' 'p' 'p' 'p' ... 'p' 'p' 'p' 'p'
array(['p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p'], dtype='<U1') - masked(acq_date)<U1'u' 'u' 'u' 'u' ... 'u' 'u' 'u' 'u'
array(['u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u'], dtype='<U1') - filtered(acq_date)<U1'n' 'n' 'n' 'n' ... 'n' 'n' 'n' 'n'
array(['n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n'], dtype='<U1') - area(acq_date)<U1'e' 'e' 'e' 'e' ... 'e' 'e' 'e' 'e'
array(['e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e'], dtype='<U1') - product_id(acq_date)<U4'1424' 'ABA0' ... '5BAD' 'BD36'
array(['1424', 'ABA0', 'E61E', 'E5B6', '5569', '71C9', 'B942', 'DCE1', '6581', '1DFD', '01B2', '9849', 'CFFC', '57F2', 'D4E6', 'B7B0', '3770', '52BB', '0579', 'AA68', '24B8', '5FF4', '5F4E', '199D', '0031', 'D082', 'E74B', 'F7ED', '65E0', '528F', '51E7', 'F5AA', 'AC38', '772E', 'F0A6', '066E', '5844', '8CD8', '6340', 'D35A', '056C', '6E6F', '0CAD', '6270', '8A4F', 'BEAB', '54B1', '8F7B', 'F9A9', 'AA59', '57E1', '81EB', '250E', '3C39', '3A2A', '9331', 'C14C', 'F1DE', '106F', 'FD00', '5D96', '971C', 'F573', 'C890', '68EA', '83D7', 'E113', 'D846', 'F4B3', 'BB26', '485C', '6683', '6153', '832B', '7418', '378D', '33F5', '1139', 'B661', '9A2F', '6943', '5BAD', 'BD36'], dtype='<U4') - acq_hour(acq_date)int6412 12 0 12 0 12 0 ... 12 12 0 0 0 0
array([12, 12, 0, 12, 0, 12, 0, 12, 0, 12, 0, 12, 12, 12, 0, 0, 12, 0, 12, 12, 12, 12, 0, 0, 12, 0, 12, 12, 12, 12, 12, 12, 0, 12, 12, 12, 12, 0, 12, 0, 12, 12, 12, 12, 12, 12, 0, 12, 12, 0, 12, 12, 12, 12, 12, 0, 12, 12, 0, 12, 12, 12, 12, 12, 0, 12, 12, 0, 0, 0, 12, 12, 12, 12, 12, 12, 12, 12, 12, 0, 0, 0, 0]) - orbital_dir(acq_date)<U4'desc' 'desc' 'asc' ... 'asc' 'asc'
array(['desc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'asc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'asc', 'asc'], dtype='<U4') - time(time)datetime64[ns]2021-06-02T12:05:57.441074 ... 2...
array(['2021-06-02T12:05:57.441074000', '2021-06-07T12:14:05.058343000', '2021-06-10T00:03:23.870450000', '2021-06-14T12:05:58.373830000', '2021-06-19T12:14:05.865662000', '2021-06-22T00:03:24.594621000', '2021-06-26T12:05:58.982444000', '2021-07-01T12:14:06.572473000', '2021-07-04T00:03:25.347747000', '2021-07-08T12:05:59.613554000', '2021-07-16T00:03:26.093195000', '2021-07-20T12:06:00.428473000', '2021-07-25T12:14:07.814919000', '2021-07-28T00:03:26.667023000', '2021-08-01T12:06:01.150857000', '2021-08-06T12:14:08.698596000', '2021-08-09T00:03:27.454588000', '2021-08-13T12:06:01.713793000', '2021-08-18T12:14:09.226637000', '2021-08-21T00:03:28.059716000', '2021-08-25T12:06:02.421490000', '2021-09-02T00:03:28.464707000', '2021-09-06T12:06:02.971474000', '2021-09-11T12:14:10.406593000', '2021-09-14T00:03:29.135941000', '2021-09-18T12:06:03.415938000', '2021-09-23T12:14:10.788780000', '2021-09-26T00:03:29.558327000', '2021-09-30T12:06:03.760613000', '2021-10-08T00:03:29.713275000', '2021-10-12T12:06:03.850150000', '2021-10-17T12:14:11.200359000', '2021-10-20T00:03:29.919736000', '2021-10-24T12:06:03.954256000', '2021-10-29T12:14:11.052769000', '2021-11-01T00:03:29.702246000', '2021-11-05T12:06:03.645002000', '2021-11-10T12:14:10.742983000', '2021-11-13T00:03:29.481869000', '2021-11-17T12:06:03.560526000', '2021-11-22T12:14:10.576899000', '2021-11-25T00:03:29.120003000', '2021-11-29T12:06:03.035677000', '2021-12-04T12:14:10.007158000', '2021-12-07T00:03:28.568419000', '2021-12-11T12:06:02.513966000', '2021-12-16T12:14:09.596874000', '2021-12-19T00:03:28.141449000', '2021-12-23T12:06:01.793721000', '2021-12-28T12:14:08.858303000', '2021-12-31T00:03:27.432317000', '2022-01-04T12:06:01.280484000', '2022-01-09T12:14:08.415805000', '2022-01-12T00:03:27.070182000', '2022-01-16T12:06:00.877035000', '2022-01-21T12:14:07.716165000', '2022-01-24T00:03:26.424832000', '2022-01-28T12:06:00.352862000', '2022-02-02T12:14:07.057390000', '2022-02-14T12:14:07.178103000', '2022-02-17T00:03:25.677942000', '2022-02-21T12:05:59.939806000', '2022-02-26T12:14:07.075742000', '2022-03-01T00:03:25.589986000', '2022-03-05T12:05:59.677073000', '2022-03-10T12:14:06.994606000', '2022-03-13T00:03:25.744897000', '2022-03-17T12:05:59.706124000', '2022-03-22T12:14:07.362180000', '2022-03-25T00:03:26.139523000', '2022-03-29T12:06:00.140262000', '2022-04-03T12:14:07.585924000', '2022-04-06T00:03:26.181999000', '2022-04-10T12:06:00.398442000', '2022-04-15T12:14:07.711367000', '2022-04-18T00:03:26.603587000', '2022-04-22T12:06:01.051309000', '2022-04-27T12:14:08.426000000', '2022-04-30T00:03:27.201903000', '2022-05-04T12:06:01.122366000', '2022-05-09T12:14:08.980398000', '2022-05-16T12:06:02.314833000', '2022-05-21T12:14:09.786375000'], dtype='datetime64[ns]') - id(time)<U66'S1A_IW_GRDH_1SDV_20210602T12054...
array(['S1A_IW_GRDH_1SDV_20210602T120544_20210602T120609_038161_0480FD_rtc', 'S1A_IW_GRDH_1SDV_20210607T121352_20210607T121417_038234_048318_rtc', 'S1A_IW_GRDH_1SDV_20210610T000311_20210610T000336_038270_04841E_rtc', 'S1A_IW_GRDH_1SDV_20210614T120545_20210614T120610_038336_04862F_rtc', 'S1A_IW_GRDH_1SDV_20210619T121353_20210619T121418_038409_04884C_rtc', 'S1A_IW_GRDH_1SDV_20210622T000312_20210622T000337_038445_04895A_rtc', 'S1A_IW_GRDH_1SDV_20210626T120546_20210626T120611_038511_048B6C_rtc', 'S1A_IW_GRDH_1SDV_20210701T121354_20210701T121419_038584_048D87_rtc', 'S1A_IW_GRDH_1SDV_20210704T000312_20210704T000337_038620_048E99_rtc', 'S1A_IW_GRDH_1SDV_20210708T120547_20210708T120612_038686_0490AD_rtc', 'S1A_IW_GRDH_1SDV_20210716T000313_20210716T000338_038795_0493DB_rtc', 'S1A_IW_GRDH_1SDV_20210720T120547_20210720T120612_038861_0495EC_rtc', 'S1A_IW_GRDH_1SDV_20210725T121355_20210725T121420_038934_04980F_rtc', 'S1A_IW_GRDH_1SDV_20210728T000314_20210728T000339_038970_04991E_rtc', 'S1A_IW_GRDH_1SDV_20210801T120548_20210801T120613_039036_049B1F_rtc', 'S1A_IW_GRDH_1SDV_20210806T121356_20210806T121421_039109_049D70_rtc', 'S1A_IW_GRDH_1SDV_20210809T000314_20210809T000339_039145_049EAC_rtc', 'S1A_IW_GRDH_1SDV_20210813T120549_20210813T120614_039211_04A0FF_rtc', 'S1A_IW_GRDH_1SDV_20210818T121356_20210818T121421_039284_04A383_rtc', 'S1A_IW_GRDH_1SDV_20210821T000315_20210821T000340_039320_04A4B9_rtc', ... 'S1A_IW_GRDH_1SDV_20220305T120547_20220305T120612_042186_0506F7_rtc', 'S1A_IW_GRDH_1SDV_20220310T121354_20220310T121419_042259_050962_rtc', 'S1A_IW_GRDH_1SDV_20220313T000313_20220313T000338_042295_050AA2_rtc', 'S1A_IW_GRDH_1SDV_20220317T120547_20220317T120612_042361_050CE5_rtc', 'S1A_IW_GRDH_1SDV_20220322T121354_20220322T121419_042434_050F59_rtc', 'S1A_IW_GRDH_1SDV_20220325T000313_20220325T000338_042470_051096_rtc', 'S1A_IW_GRDH_1SDV_20220329T120547_20220329T120612_042536_0512D7_rtc', 'S1A_IW_GRDH_1SDV_20220403T121355_20220403T121420_042609_05154A_rtc', 'S1A_IW_GRDH_1SDV_20220406T000313_20220406T000338_042645_051682_rtc', 'S1A_IW_GRDH_1SDV_20220410T120547_20220410T120612_042711_0518C0_rtc', 'S1A_IW_GRDH_1SDV_20220415T121355_20220415T121420_042784_051B30_rtc', 'S1A_IW_GRDH_1SDV_20220418T000314_20220418T000339_042820_051C63_rtc', 'S1A_IW_GRDH_1SDV_20220422T120548_20220422T120613_042886_051E96_rtc', 'S1A_IW_GRDH_1SDV_20220427T121355_20220427T121420_042959_0520EE_rtc', 'S1A_IW_GRDH_1SDV_20220430T000314_20220430T000339_042995_05221E_rtc', 'S1A_IW_GRDH_1SDV_20220504T120548_20220504T120613_043061_05245F_rtc', 'S1A_IW_GRDH_1SDV_20220509T121356_20220509T121421_043134_0526C4_rtc', 'S1A_IW_GRDH_1SDV_20220516T120549_20220516T120614_043236_0529DD_rtc', 'S1A_IW_GRDH_1SDV_20220521T121357_20220521T121422_043309_052C00_rtc'], dtype='<U66') - band(band)<U2'vh' 'vv'
array(['vh', 'vv'], dtype='<U2')
- sar:looks_azimuth()int641
array(1)
- sat:relative_orbit(time)int64114 12 48 114 12 ... 114 12 114 12
array([114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 114, 12]) - s1:instrument_configuration_ID(time)<U1'6' '6' '6' '6' ... '7' '7' '7' '7'
array(['6', '6', '6', '6', '6', '6', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7'], dtype='<U1') - s1:orbit_source(time)<U8'DOWNLINK' 'DOWNLINK' ... 'RESORB'
array(['DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB'], dtype='<U8') - end_datetime(time)<U32'2021-06-02 12:06:09.940813+00:0...
array(['2021-06-02 12:06:09.940813+00:00', '2021-06-07 12:14:17.557268+00:00', '2021-06-10 00:03:36.369821+00:00', '2021-06-14 12:06:10.872859+00:00', '2021-06-19 12:14:18.365410+00:00', '2021-06-22 00:03:37.093898+00:00', '2021-06-26 12:06:11.482149+00:00', '2021-07-01 12:14:19.071405+00:00', '2021-07-04 00:03:37.847082+00:00', '2021-07-08 12:06:12.112559+00:00', '2021-07-16 00:03:38.592472+00:00', '2021-07-20 12:06:12.927516+00:00', '2021-07-25 12:14:20.313830+00:00', '2021-07-28 00:03:39.166376+00:00', '2021-08-01 12:06:13.649807+00:00', '2021-08-06 12:14:21.197519+00:00', '2021-08-09 00:03:39.953924+00:00', '2021-08-13 12:06:14.213502+00:00', '2021-08-18 12:14:21.725577+00:00', '2021-08-21 00:03:40.559065+00:00', ... '2022-03-01 00:03:38.089393+00:00', '2022-03-05 12:06:12.176227+00:00', '2022-03-10 12:14:19.493699+00:00', '2022-03-13 00:03:38.244245+00:00', '2022-03-17 12:06:12.205265+00:00', '2022-03-22 12:14:19.861261+00:00', '2022-03-25 00:03:38.638828+00:00', '2022-03-29 12:06:12.640119+00:00', '2022-04-03 12:14:20.084951+00:00', '2022-04-06 00:03:38.681256+00:00', '2022-04-10 12:06:12.897487+00:00', '2022-04-15 12:14:20.210307+00:00', '2022-04-18 00:03:39.102936+00:00', '2022-04-22 12:06:13.551041+00:00', '2022-04-27 12:14:20.924939+00:00', '2022-04-30 00:03:39.701264+00:00', '2022-05-04 12:06:13.622093+00:00', '2022-05-09 12:14:21.479341+00:00', '2022-05-16 12:06:14.814536+00:00', '2022-05-21 12:14:22.285304+00:00'], dtype='<U32') - sat:orbit_state(time)<U10'ascending' ... 'ascending'
array(['ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'ascending', 'ascending'], dtype='<U10') - platform()<U11'SENTINEL-1A'
array('SENTINEL-1A', dtype='<U11') - s1:slice_number(time)<U2'6' '6' '12' '6' ... '6' '6' '6'
array(['6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '6', '6'], dtype='<U2') - start_datetime(time)<U32'2021-06-02 12:05:44.941334+00:0...
array(['2021-06-02 12:05:44.941334+00:00', '2021-06-07 12:13:52.559418+00:00', '2021-06-10 00:03:11.371078+00:00', '2021-06-14 12:05:45.874802+00:00', '2021-06-19 12:13:53.365914+00:00', '2021-06-22 00:03:12.095344+00:00', '2021-06-26 12:05:46.482738+00:00', '2021-07-01 12:13:54.073541+00:00', '2021-07-04 00:03:12.848412+00:00', '2021-07-08 12:05:47.114550+00:00', '2021-07-16 00:03:13.593918+00:00', '2021-07-20 12:05:47.929430+00:00', '2021-07-25 12:13:55.316007+00:00', '2021-07-28 00:03:14.167671+00:00', '2021-08-01 12:05:48.651907+00:00', '2021-08-06 12:13:56.199672+00:00', '2021-08-09 00:03:14.955252+00:00', '2021-08-13 12:05:49.214084+00:00', '2021-08-18 12:13:56.727697+00:00', '2021-08-21 00:03:15.560368+00:00', ... '2022-03-01 00:03:13.090580+00:00', '2022-03-05 12:05:47.177919+00:00', '2022-03-10 12:13:54.495512+00:00', '2022-03-13 00:03:13.245549+00:00', '2022-03-17 12:05:47.206984+00:00', '2022-03-22 12:13:54.863098+00:00', '2022-03-25 00:03:13.640217+00:00', '2022-03-29 12:05:47.640405+00:00', '2022-04-03 12:13:55.086898+00:00', '2022-04-06 00:03:13.682743+00:00', '2022-04-10 12:05:47.899396+00:00', '2022-04-15 12:13:55.212427+00:00', '2022-04-18 00:03:14.104239+00:00', '2022-04-22 12:05:48.551577+00:00', '2022-04-27 12:13:55.927061+00:00', '2022-04-30 00:03:14.702542+00:00', '2022-05-04 12:05:48.622638+00:00', '2022-05-09 12:13:56.481455+00:00', '2022-05-16 12:05:49.815129+00:00', '2022-05-21 12:13:57.287447+00:00'], dtype='<U32') - sar:resolution_azimuth()int6422
array(22)
- s1:resolution()<U4'high'
array('high', dtype='<U4') - sar:frequency_band()<U1'C'
array('C', dtype='<U1') - sar:center_frequency()float645.405
array(5.405)
- sar:pixel_spacing_azimuth()int6410
array(10)
- sat:platform_international_designator()<U9'2014-016A'
array('2014-016A', dtype='<U9') - constellation()<U10'Sentinel-1'
array('Sentinel-1', dtype='<U10') - sar:polarizations()object{'VV', 'VH'}
array({'VV', 'VH'}, dtype=object) - proj:epsg()int6432645
array(32645)
- sar:pixel_spacing_range()int6410
array(10)
- sar:looks_range()int645
array(5)
- sar:resolution_range()int6420
array(20)
- sat:absolute_orbit(time)int6438161 38234 38270 ... 43236 43309
array([38161, 38234, 38270, 38336, 38409, 38445, 38511, 38584, 38620, 38686, 38795, 38861, 38934, 38970, 39036, 39109, 39145, 39211, 39284, 39320, 39386, 39495, 39561, 39634, 39670, 39736, 39809, 39845, 39911, 40020, 40086, 40159, 40195, 40261, 40334, 40370, 40436, 40509, 40545, 40611, 40684, 40720, 40786, 40859, 40895, 40961, 41034, 41070, 41136, 41209, 41245, 41311, 41384, 41420, 41486, 41559, 41595, 41661, 41734, 41909, 41945, 42011, 42084, 42120, 42186, 42259, 42295, 42361, 42434, 42470, 42536, 42609, 42645, 42711, 42784, 42820, 42886, 42959, 42995, 43061, 43134, 43236, 43309]) - sar:looks_equivalent_number()float644.4
array(4.4)
- sar:observation_direction()<U5'right'
array('right', dtype='<U5') - sar:product_type()<U3'GRD'
array('GRD', dtype='<U3') - s1:product_timeliness()<U8'Fast-24h'
array('Fast-24h', dtype='<U8') - sar:instrument_mode()<U2'IW'
array('IW', dtype='<U2') - s1:datatake_id(time)<U6'295165' '295704' ... '338944'
array(['295165', '295704', '295966', '296495', '297036', '297306', '297836', '298375', '298649', '299181', '299995', '300524', '301071', '301342', '301855', '302448', '302764', '303359', '304003', '304313', '304892', '305843', '306445', '307077', '307396', '307963', '308582', '308907', '309503', '310456', '311058', '311701', '312022', '312622', '313256', '313577', '314182', '314824', '315135', '315727', '316364', '316686', '317281', '317928', '318241', '318833', '319453', '319760', '320320', '320939', '321239', '321841', '322459', '322765', '323304', '323926', '324244', '324822', '325460', '327023', '327346', '327944', '328566', '328879', '329463', '330082', '330402', '330981', '331609', '331926', '332503', '333130', '333442', '334016', '334640', '334947', '335510', '336110', '336414', '336991', '337604', '338397', '338944'], dtype='<U6') - s1:total_slices(time)<U2'21' '20' '17' ... '20' '21' '20'
array(['21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '21', '20'], dtype='<U2') - s1:processing_level()<U1'1'
array('1', dtype='<U1') - title(band)<U41'VH: vertical transmit, horizont...
array(['VH: vertical transmit, horizontal receive', 'VV: vertical transmit, vertical receive'], dtype='<U41') - description(band)<U173'Terrain-corrected gamma naught ...
array(['Terrain-corrected gamma naught values of signal transmitted with vertical polarization and received with horizontal polarization with radiometric terrain correction applied.', 'Terrain-corrected gamma naught values of signal transmitted with vertical polarization and received with vertical polarization with radiometric terrain correction applied.'], dtype='<U173') - raster:bands()object{'nodata': -32768, 'data_type': ...
array({'nodata': -32768, 'data_type': 'float32', 'spatial_resolution': 10.0}, dtype=object) - epsg()int6432645
array(32645)
- vv(acq_date, y, x, time, band)float64dask.array<chunksize=(1, 396, 290, 1, 1), meta=np.ndarray>
Array Chunk Bytes 11.79 GiB 897.19 kiB Shape (83, 396, 290, 83, 2) (1, 396, 290, 1, 1) Count 42 Graph Layers 13778 Chunks Type float64 numpy.ndarray - vh(acq_date, y, x, time, band)float64dask.array<chunksize=(1, 396, 290, 1, 1), meta=np.ndarray>
Array Chunk Bytes 11.79 GiB 897.19 kiB Shape (83, 396, 290, 83, 2) (1, 396, 290, 1, 1) Count 53 Graph Layers 13778 Chunks Type float64 numpy.ndarray - ls(acq_date, y, x, time, band)float64dask.array<chunksize=(1, 396, 290, 1, 1), meta=np.ndarray>
Array Chunk Bytes 11.79 GiB 897.19 kiB Shape (83, 396, 290, 83, 2) (1, 396, 290, 1, 1) Count 57 Graph Layers 13778 Chunks Type float64 numpy.ndarray
Oops, this isn’t what we want. Now we have a 5-dimensional object instead of a 4-dimensional object, because the dimensions of the asf_subset object were broadcast onto the pc_downsample object, meaning that we have both a time and a acq_date object.
Let’s go back and try to fix this…
pc_downsample.isel(time=0).time.values
numpy.datetime64('2021-06-02T12:05:57.441074000')
asf_subset = asf_subset.rename({'acq_date':'time'})
asf_subset.isel(time=0).time.values
numpy.datetime64('2021-06-02T00:00:00.000000000')
Okay, so we need to take the time off of the pc_downsample time coordinates:
pc_downsample.coords['time'] = pc_downsample.time.dt.date
pc_downsample.coords.dtypes
Frozen({'time': dtype('O'), 'id': dtype('<U66'), 'band': dtype('<U2'), 'sar:looks_azimuth': dtype('int64'), 'sat:relative_orbit': dtype('int64'), 's1:instrument_configuration_ID': dtype('<U1'), 's1:orbit_source': dtype('<U8'), 'end_datetime': dtype('<U32'), 'sat:orbit_state': dtype('<U10'), 'platform': dtype('<U11'), 's1:slice_number': dtype('<U2'), 'start_datetime': dtype('<U32'), 'sar:resolution_azimuth': dtype('int64'), 's1:resolution': dtype('<U4'), 'sar:frequency_band': dtype('<U1'), 'sar:center_frequency': dtype('float64'), 'sar:pixel_spacing_azimuth': dtype('int64'), 'sat:platform_international_designator': dtype('<U9'), 'constellation': dtype('<U10'), 'sar:polarizations': dtype('O'), 'proj:epsg': dtype('int64'), 'sar:pixel_spacing_range': dtype('int64'), 'sar:looks_range': dtype('int64'), 'sar:resolution_range': dtype('int64'), 'sat:absolute_orbit': dtype('int64'), 'sar:looks_equivalent_number': dtype('float64'), 'sar:observation_direction': dtype('<U5'), 'sar:product_type': dtype('<U3'), 's1:product_timeliness': dtype('<U8'), 'sar:instrument_mode': dtype('<U2'), 's1:datatake_id': dtype('<U6'), 's1:total_slices': dtype('<U2'), 's1:processing_level': dtype('<U1'), 'title': dtype('<U41'), 'description': dtype('<U173'), 'raster:bands': dtype('O'), 'epsg': dtype('int64'), 'granule_id': dtype('<U62'), 'data_take_id': dtype('<U6'), 'x': dtype('float64'), 'y': dtype('float64')})
asf_subset.coords.dtypes
Frozen({'time': dtype('<M8[ns]'), 'granule_id': dtype('<U67'), 'x': dtype('float64'), 'y': dtype('float64'), 'spatial_ref': dtype('int64'), 'sensor': dtype('<U3'), 'beam_mode': dtype('<U2'), 'acquisition_time': dtype('<M8[ns]'), 'polarisation_type': dtype('<U2'), 'orbit_type': dtype('<U1'), 'terrain_correction_pixel_spacing': dtype('<U5'), 'output_format': dtype('<U1'), 'output_type': dtype('<U1'), 'masked': dtype('<U1'), 'filtered': dtype('<U1'), 'area': dtype('<U1'), 'product_id': dtype('<U4'), 'acq_hour': dtype('int64'), 'orbital_dir': dtype('<U4'), 'data_take_id': dtype('<U6')})
Now let’s try again:
pc_mask = xr.where(asf_subset.notnull(), pc_downsample,np.nan)
pc_mask
<xarray.Dataset>
Dimensions: (time: 83, y: 396, x: 290, band: 2)
Coordinates: (12/55)
* time (time) datetime64[ns] 2021-06-02 ....
* x (x) float64 6.194e+05 ... 6.281e+05
* y (y) float64 3.102e+06 ... 3.09e+06
spatial_ref int64 0
sensor (time) <U3 'S1A' 'S1A' ... 'S1A'
beam_mode (time) <U2 'IW' 'IW' ... 'IW' 'IW'
... ...
s1:total_slices (time) <U2 '21' '20' ... '21' '20'
s1:processing_level <U1 '1'
title (band) <U41 'VH: vertical transmit...
description (band) <U173 'Terrain-corrected ga...
raster:bands object {'nodata': -32768, 'data_ty...
epsg int64 32645
Data variables:
vv (time, y, x, band) float64 dask.array<chunksize=(1, 396, 290, 1), meta=np.ndarray>
vh (time, y, x, band) float64 dask.array<chunksize=(1, 396, 290, 1), meta=np.ndarray>
ls (time, y, x, band) float64 dask.array<chunksize=(1, 396, 290, 1), meta=np.ndarray>- time: 83
- y: 396
- x: 290
- band: 2
- time(time)datetime64[ns]2021-06-02 ... 2022-05-21
array(['2021-06-02T00:00:00.000000000', '2021-06-07T00:00:00.000000000', '2021-06-10T00:00:00.000000000', '2021-06-14T00:00:00.000000000', '2021-06-19T00:00:00.000000000', '2021-06-22T00:00:00.000000000', '2021-06-26T00:00:00.000000000', '2021-07-01T00:00:00.000000000', '2021-07-04T00:00:00.000000000', '2021-07-08T00:00:00.000000000', '2021-07-16T00:00:00.000000000', '2021-07-20T00:00:00.000000000', '2021-07-25T00:00:00.000000000', '2021-07-28T00:00:00.000000000', '2021-08-01T00:00:00.000000000', '2021-08-06T00:00:00.000000000', '2021-08-09T00:00:00.000000000', '2021-08-13T00:00:00.000000000', '2021-08-18T00:00:00.000000000', '2021-08-21T00:00:00.000000000', '2021-08-25T00:00:00.000000000', '2021-09-02T00:00:00.000000000', '2021-09-06T00:00:00.000000000', '2021-09-11T00:00:00.000000000', '2021-09-14T00:00:00.000000000', '2021-09-18T00:00:00.000000000', '2021-09-23T00:00:00.000000000', '2021-09-26T00:00:00.000000000', '2021-09-30T00:00:00.000000000', '2021-10-08T00:00:00.000000000', '2021-10-12T00:00:00.000000000', '2021-10-17T00:00:00.000000000', '2021-10-20T00:00:00.000000000', '2021-10-24T00:00:00.000000000', '2021-10-29T00:00:00.000000000', '2021-11-01T00:00:00.000000000', '2021-11-05T00:00:00.000000000', '2021-11-10T00:00:00.000000000', '2021-11-13T00:00:00.000000000', '2021-11-17T00:00:00.000000000', '2021-11-22T00:00:00.000000000', '2021-11-25T00:00:00.000000000', '2021-11-29T00:00:00.000000000', '2021-12-04T00:00:00.000000000', '2021-12-07T00:00:00.000000000', '2021-12-11T00:00:00.000000000', '2021-12-16T00:00:00.000000000', '2021-12-19T00:00:00.000000000', '2021-12-23T00:00:00.000000000', '2021-12-28T00:00:00.000000000', '2021-12-31T00:00:00.000000000', '2022-01-04T00:00:00.000000000', '2022-01-09T00:00:00.000000000', '2022-01-12T00:00:00.000000000', '2022-01-16T00:00:00.000000000', '2022-01-21T00:00:00.000000000', '2022-01-24T00:00:00.000000000', '2022-01-28T00:00:00.000000000', '2022-02-02T00:00:00.000000000', '2022-02-14T00:00:00.000000000', '2022-02-17T00:00:00.000000000', '2022-02-21T00:00:00.000000000', '2022-02-26T00:00:00.000000000', '2022-03-01T00:00:00.000000000', '2022-03-05T00:00:00.000000000', '2022-03-10T00:00:00.000000000', '2022-03-13T00:00:00.000000000', '2022-03-17T00:00:00.000000000', '2022-03-22T00:00:00.000000000', '2022-03-25T00:00:00.000000000', '2022-03-29T00:00:00.000000000', '2022-04-03T00:00:00.000000000', '2022-04-06T00:00:00.000000000', '2022-04-10T00:00:00.000000000', '2022-04-15T00:00:00.000000000', '2022-04-18T00:00:00.000000000', '2022-04-22T00:00:00.000000000', '2022-04-27T00:00:00.000000000', '2022-04-30T00:00:00.000000000', '2022-05-04T00:00:00.000000000', '2022-05-09T00:00:00.000000000', '2022-05-16T00:00:00.000000000', '2022-05-21T00:00:00.000000000'], dtype='datetime64[ns]') - x(x)float646.194e+05 6.195e+05 ... 6.281e+05
array([619425., 619455., 619485., ..., 628035., 628065., 628095.])
- y(y)float643.102e+06 3.102e+06 ... 3.09e+06
array([3101655., 3101625., 3101595., ..., 3089865., 3089835., 3089805.])
- spatial_ref()int640
array(0)
- sensor(time)<U3'S1A' 'S1A' 'S1A' ... 'S1A' 'S1A'
array(['S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A'], dtype='<U3') - beam_mode(time)<U2'IW' 'IW' 'IW' ... 'IW' 'IW' 'IW'
array(['IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW'], dtype='<U2') - acquisition_time(time)datetime64[ns]2022-10-21T12:14:14 ... 2022-10-...
array(['2022-10-21T12:14:14.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:14:20.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T12:05:43.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:05:43.000000000', '2022-10-21T00:03:14.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:52.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T12:13:50.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T12:14:17.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T00:03:10.000000000', '2022-10-21T12:05:45.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:05:50.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:52.000000000', '2022-10-21T00:03:08.000000000', ... '2022-10-21T12:05:42.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:07.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:50.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T00:03:14.000000000', '2022-10-21T12:05:44.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:13:49.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:45.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T00:03:16.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T12:13:51.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T00:03:09.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T00:03:12.000000000'], dtype='datetime64[ns]') - polarisation_type(time)<U2'DV' 'DV' 'DV' ... 'DV' 'DV' 'DV'
array(['DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV'], dtype='<U2') - orbit_type(time)<U1'P' 'P' 'P' 'P' ... 'P' 'P' 'P' 'P'
array(['P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P'], dtype='<U1') - terrain_correction_pixel_spacing(time)<U5'RTC30' 'RTC30' ... 'RTC30' 'RTC30'
array(['RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30'], dtype='<U5') - output_format(time)<U1'g' 'g' 'g' 'g' ... 'g' 'g' 'g' 'g'
array(['g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g'], dtype='<U1') - output_type(time)<U1'p' 'p' 'p' 'p' ... 'p' 'p' 'p' 'p'
array(['p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p'], dtype='<U1') - masked(time)<U1'u' 'u' 'u' 'u' ... 'u' 'u' 'u' 'u'
array(['u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u'], dtype='<U1') - filtered(time)<U1'n' 'n' 'n' 'n' ... 'n' 'n' 'n' 'n'
array(['n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n'], dtype='<U1') - area(time)<U1'e' 'e' 'e' 'e' ... 'e' 'e' 'e' 'e'
array(['e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e'], dtype='<U1') - product_id(time)<U4'1424' 'ABA0' ... '5BAD' 'BD36'
array(['1424', 'ABA0', 'E61E', 'E5B6', '5569', '71C9', 'B942', 'DCE1', '6581', '1DFD', '01B2', '9849', 'CFFC', '57F2', 'D4E6', 'B7B0', '3770', '52BB', '0579', 'AA68', '24B8', '5FF4', '5F4E', '199D', '0031', 'D082', 'E74B', 'F7ED', '65E0', '528F', '51E7', 'F5AA', 'AC38', '772E', 'F0A6', '066E', '5844', '8CD8', '6340', 'D35A', '056C', '6E6F', '0CAD', '6270', '8A4F', 'BEAB', '54B1', '8F7B', 'F9A9', 'AA59', '57E1', '81EB', '250E', '3C39', '3A2A', '9331', 'C14C', 'F1DE', '106F', 'FD00', '5D96', '971C', 'F573', 'C890', '68EA', '83D7', 'E113', 'D846', 'F4B3', 'BB26', '485C', '6683', '6153', '832B', '7418', '378D', '33F5', '1139', 'B661', '9A2F', '6943', '5BAD', 'BD36'], dtype='<U4') - acq_hour(time)int6412 12 0 12 0 12 0 ... 12 12 0 0 0 0
array([12, 12, 0, 12, 0, 12, 0, 12, 0, 12, 0, 12, 12, 12, 0, 0, 12, 0, 12, 12, 12, 12, 0, 0, 12, 0, 12, 12, 12, 12, 12, 12, 0, 12, 12, 12, 12, 0, 12, 0, 12, 12, 12, 12, 12, 12, 0, 12, 12, 0, 12, 12, 12, 12, 12, 0, 12, 12, 0, 12, 12, 12, 12, 12, 0, 12, 12, 0, 0, 0, 12, 12, 12, 12, 12, 12, 12, 12, 12, 0, 0, 0, 0]) - orbital_dir(time)<U4'desc' 'desc' 'asc' ... 'asc' 'asc'
array(['desc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'asc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'asc', 'asc'], dtype='<U4') - data_take_id(time)<U6'0480FD' '048318' ... '052C00'
array(['0480FD', '048318', '04841E', '04862F', '04884C', '04895A', '048B6C', '048D87', '048E99', '0490AD', '0493DB', '0495EC', '04980F', '04991E', '049B1F', '049D70', '049EAC', '04A0FF', '04A383', '04A4B9', '04A6FC', '04AAB3', '04AD0D', '04AF85', '04B0C4', '04B2FB', '04B566', '04B6AB', '04B8FF', '04BCB8', '04BF12', '04C195', '04C2D6', '04C52E', '04C7A8', '04C8E9', '04CB46', '04CDC8', '04CEFF', '04D14F', '04D3CC', '04D50E', '04D761', '04D9E8', '04DB21', '04DD71', '04DFDD', '04E110', '04E340', '04E5AB', '04E6D7', '04E931', '04EB9B', '04ECCD', '04EEE8', '04F156', '04F294', '04F4D6', '04F754', '04FD6F', '04FEB2', '050108', '050376', '0504AF', '0506F7', '050962', '050AA2', '050CE5', '050F59', '051096', '0512D7', '05154A', '051682', '0518C0', '051B30', '051C63', '051E96', '0520EE', '05221E', '05245F', '0526C4', '0529DD', '052C00'], dtype='<U6') - id(time)<U66'S1A_IW_GRDH_1SDV_20210602T12054...
array(['S1A_IW_GRDH_1SDV_20210602T120544_20210602T120609_038161_0480FD_rtc', 'S1A_IW_GRDH_1SDV_20210607T121352_20210607T121417_038234_048318_rtc', 'S1A_IW_GRDH_1SDV_20210610T000311_20210610T000336_038270_04841E_rtc', 'S1A_IW_GRDH_1SDV_20210614T120545_20210614T120610_038336_04862F_rtc', 'S1A_IW_GRDH_1SDV_20210619T121353_20210619T121418_038409_04884C_rtc', 'S1A_IW_GRDH_1SDV_20210622T000312_20210622T000337_038445_04895A_rtc', 'S1A_IW_GRDH_1SDV_20210626T120546_20210626T120611_038511_048B6C_rtc', 'S1A_IW_GRDH_1SDV_20210701T121354_20210701T121419_038584_048D87_rtc', 'S1A_IW_GRDH_1SDV_20210704T000312_20210704T000337_038620_048E99_rtc', 'S1A_IW_GRDH_1SDV_20210708T120547_20210708T120612_038686_0490AD_rtc', 'S1A_IW_GRDH_1SDV_20210716T000313_20210716T000338_038795_0493DB_rtc', 'S1A_IW_GRDH_1SDV_20210720T120547_20210720T120612_038861_0495EC_rtc', 'S1A_IW_GRDH_1SDV_20210725T121355_20210725T121420_038934_04980F_rtc', 'S1A_IW_GRDH_1SDV_20210728T000314_20210728T000339_038970_04991E_rtc', 'S1A_IW_GRDH_1SDV_20210801T120548_20210801T120613_039036_049B1F_rtc', 'S1A_IW_GRDH_1SDV_20210806T121356_20210806T121421_039109_049D70_rtc', 'S1A_IW_GRDH_1SDV_20210809T000314_20210809T000339_039145_049EAC_rtc', 'S1A_IW_GRDH_1SDV_20210813T120549_20210813T120614_039211_04A0FF_rtc', 'S1A_IW_GRDH_1SDV_20210818T121356_20210818T121421_039284_04A383_rtc', 'S1A_IW_GRDH_1SDV_20210821T000315_20210821T000340_039320_04A4B9_rtc', ... 'S1A_IW_GRDH_1SDV_20220305T120547_20220305T120612_042186_0506F7_rtc', 'S1A_IW_GRDH_1SDV_20220310T121354_20220310T121419_042259_050962_rtc', 'S1A_IW_GRDH_1SDV_20220313T000313_20220313T000338_042295_050AA2_rtc', 'S1A_IW_GRDH_1SDV_20220317T120547_20220317T120612_042361_050CE5_rtc', 'S1A_IW_GRDH_1SDV_20220322T121354_20220322T121419_042434_050F59_rtc', 'S1A_IW_GRDH_1SDV_20220325T000313_20220325T000338_042470_051096_rtc', 'S1A_IW_GRDH_1SDV_20220329T120547_20220329T120612_042536_0512D7_rtc', 'S1A_IW_GRDH_1SDV_20220403T121355_20220403T121420_042609_05154A_rtc', 'S1A_IW_GRDH_1SDV_20220406T000313_20220406T000338_042645_051682_rtc', 'S1A_IW_GRDH_1SDV_20220410T120547_20220410T120612_042711_0518C0_rtc', 'S1A_IW_GRDH_1SDV_20220415T121355_20220415T121420_042784_051B30_rtc', 'S1A_IW_GRDH_1SDV_20220418T000314_20220418T000339_042820_051C63_rtc', 'S1A_IW_GRDH_1SDV_20220422T120548_20220422T120613_042886_051E96_rtc', 'S1A_IW_GRDH_1SDV_20220427T121355_20220427T121420_042959_0520EE_rtc', 'S1A_IW_GRDH_1SDV_20220430T000314_20220430T000339_042995_05221E_rtc', 'S1A_IW_GRDH_1SDV_20220504T120548_20220504T120613_043061_05245F_rtc', 'S1A_IW_GRDH_1SDV_20220509T121356_20220509T121421_043134_0526C4_rtc', 'S1A_IW_GRDH_1SDV_20220516T120549_20220516T120614_043236_0529DD_rtc', 'S1A_IW_GRDH_1SDV_20220521T121357_20220521T121422_043309_052C00_rtc'], dtype='<U66') - band(band)<U2'vh' 'vv'
array(['vh', 'vv'], dtype='<U2')
- sar:looks_azimuth()int641
array(1)
- sat:relative_orbit(time)int64114 12 48 114 12 ... 114 12 114 12
array([114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 114, 12]) - s1:instrument_configuration_ID(time)<U1'6' '6' '6' '6' ... '7' '7' '7' '7'
array(['6', '6', '6', '6', '6', '6', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7'], dtype='<U1') - s1:orbit_source(time)<U8'DOWNLINK' 'DOWNLINK' ... 'RESORB'
array(['DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB'], dtype='<U8') - end_datetime(time)<U32'2021-06-02 12:06:09.940813+00:0...
array(['2021-06-02 12:06:09.940813+00:00', '2021-06-07 12:14:17.557268+00:00', '2021-06-10 00:03:36.369821+00:00', '2021-06-14 12:06:10.872859+00:00', '2021-06-19 12:14:18.365410+00:00', '2021-06-22 00:03:37.093898+00:00', '2021-06-26 12:06:11.482149+00:00', '2021-07-01 12:14:19.071405+00:00', '2021-07-04 00:03:37.847082+00:00', '2021-07-08 12:06:12.112559+00:00', '2021-07-16 00:03:38.592472+00:00', '2021-07-20 12:06:12.927516+00:00', '2021-07-25 12:14:20.313830+00:00', '2021-07-28 00:03:39.166376+00:00', '2021-08-01 12:06:13.649807+00:00', '2021-08-06 12:14:21.197519+00:00', '2021-08-09 00:03:39.953924+00:00', '2021-08-13 12:06:14.213502+00:00', '2021-08-18 12:14:21.725577+00:00', '2021-08-21 00:03:40.559065+00:00', ... '2022-03-01 00:03:38.089393+00:00', '2022-03-05 12:06:12.176227+00:00', '2022-03-10 12:14:19.493699+00:00', '2022-03-13 00:03:38.244245+00:00', '2022-03-17 12:06:12.205265+00:00', '2022-03-22 12:14:19.861261+00:00', '2022-03-25 00:03:38.638828+00:00', '2022-03-29 12:06:12.640119+00:00', '2022-04-03 12:14:20.084951+00:00', '2022-04-06 00:03:38.681256+00:00', '2022-04-10 12:06:12.897487+00:00', '2022-04-15 12:14:20.210307+00:00', '2022-04-18 00:03:39.102936+00:00', '2022-04-22 12:06:13.551041+00:00', '2022-04-27 12:14:20.924939+00:00', '2022-04-30 00:03:39.701264+00:00', '2022-05-04 12:06:13.622093+00:00', '2022-05-09 12:14:21.479341+00:00', '2022-05-16 12:06:14.814536+00:00', '2022-05-21 12:14:22.285304+00:00'], dtype='<U32') - sat:orbit_state(time)<U10'ascending' ... 'ascending'
array(['ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'ascending', 'ascending'], dtype='<U10') - platform()<U11'SENTINEL-1A'
array('SENTINEL-1A', dtype='<U11') - s1:slice_number(time)<U2'6' '6' '12' '6' ... '6' '6' '6'
array(['6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '6', '6'], dtype='<U2') - start_datetime(time)<U32'2021-06-02 12:05:44.941334+00:0...
array(['2021-06-02 12:05:44.941334+00:00', '2021-06-07 12:13:52.559418+00:00', '2021-06-10 00:03:11.371078+00:00', '2021-06-14 12:05:45.874802+00:00', '2021-06-19 12:13:53.365914+00:00', '2021-06-22 00:03:12.095344+00:00', '2021-06-26 12:05:46.482738+00:00', '2021-07-01 12:13:54.073541+00:00', '2021-07-04 00:03:12.848412+00:00', '2021-07-08 12:05:47.114550+00:00', '2021-07-16 00:03:13.593918+00:00', '2021-07-20 12:05:47.929430+00:00', '2021-07-25 12:13:55.316007+00:00', '2021-07-28 00:03:14.167671+00:00', '2021-08-01 12:05:48.651907+00:00', '2021-08-06 12:13:56.199672+00:00', '2021-08-09 00:03:14.955252+00:00', '2021-08-13 12:05:49.214084+00:00', '2021-08-18 12:13:56.727697+00:00', '2021-08-21 00:03:15.560368+00:00', ... '2022-03-01 00:03:13.090580+00:00', '2022-03-05 12:05:47.177919+00:00', '2022-03-10 12:13:54.495512+00:00', '2022-03-13 00:03:13.245549+00:00', '2022-03-17 12:05:47.206984+00:00', '2022-03-22 12:13:54.863098+00:00', '2022-03-25 00:03:13.640217+00:00', '2022-03-29 12:05:47.640405+00:00', '2022-04-03 12:13:55.086898+00:00', '2022-04-06 00:03:13.682743+00:00', '2022-04-10 12:05:47.899396+00:00', '2022-04-15 12:13:55.212427+00:00', '2022-04-18 00:03:14.104239+00:00', '2022-04-22 12:05:48.551577+00:00', '2022-04-27 12:13:55.927061+00:00', '2022-04-30 00:03:14.702542+00:00', '2022-05-04 12:05:48.622638+00:00', '2022-05-09 12:13:56.481455+00:00', '2022-05-16 12:05:49.815129+00:00', '2022-05-21 12:13:57.287447+00:00'], dtype='<U32') - sar:resolution_azimuth()int6422
array(22)
- s1:resolution()<U4'high'
array('high', dtype='<U4') - sar:frequency_band()<U1'C'
array('C', dtype='<U1') - sar:center_frequency()float645.405
array(5.405)
- sar:pixel_spacing_azimuth()int6410
array(10)
- sat:platform_international_designator()<U9'2014-016A'
array('2014-016A', dtype='<U9') - constellation()<U10'Sentinel-1'
array('Sentinel-1', dtype='<U10') - sar:polarizations()object{'VV', 'VH'}
array({'VV', 'VH'}, dtype=object) - proj:epsg()int6432645
array(32645)
- sar:pixel_spacing_range()int6410
array(10)
- sar:looks_range()int645
array(5)
- sar:resolution_range()int6420
array(20)
- sat:absolute_orbit(time)int6438161 38234 38270 ... 43236 43309
array([38161, 38234, 38270, 38336, 38409, 38445, 38511, 38584, 38620, 38686, 38795, 38861, 38934, 38970, 39036, 39109, 39145, 39211, 39284, 39320, 39386, 39495, 39561, 39634, 39670, 39736, 39809, 39845, 39911, 40020, 40086, 40159, 40195, 40261, 40334, 40370, 40436, 40509, 40545, 40611, 40684, 40720, 40786, 40859, 40895, 40961, 41034, 41070, 41136, 41209, 41245, 41311, 41384, 41420, 41486, 41559, 41595, 41661, 41734, 41909, 41945, 42011, 42084, 42120, 42186, 42259, 42295, 42361, 42434, 42470, 42536, 42609, 42645, 42711, 42784, 42820, 42886, 42959, 42995, 43061, 43134, 43236, 43309]) - sar:looks_equivalent_number()float644.4
array(4.4)
- sar:observation_direction()<U5'right'
array('right', dtype='<U5') - sar:product_type()<U3'GRD'
array('GRD', dtype='<U3') - s1:product_timeliness()<U8'Fast-24h'
array('Fast-24h', dtype='<U8') - sar:instrument_mode()<U2'IW'
array('IW', dtype='<U2') - s1:datatake_id(time)<U6'295165' '295704' ... '338944'
array(['295165', '295704', '295966', '296495', '297036', '297306', '297836', '298375', '298649', '299181', '299995', '300524', '301071', '301342', '301855', '302448', '302764', '303359', '304003', '304313', '304892', '305843', '306445', '307077', '307396', '307963', '308582', '308907', '309503', '310456', '311058', '311701', '312022', '312622', '313256', '313577', '314182', '314824', '315135', '315727', '316364', '316686', '317281', '317928', '318241', '318833', '319453', '319760', '320320', '320939', '321239', '321841', '322459', '322765', '323304', '323926', '324244', '324822', '325460', '327023', '327346', '327944', '328566', '328879', '329463', '330082', '330402', '330981', '331609', '331926', '332503', '333130', '333442', '334016', '334640', '334947', '335510', '336110', '336414', '336991', '337604', '338397', '338944'], dtype='<U6') - s1:total_slices(time)<U2'21' '20' '17' ... '20' '21' '20'
array(['21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '21', '20'], dtype='<U2') - s1:processing_level()<U1'1'
array('1', dtype='<U1') - title(band)<U41'VH: vertical transmit, horizont...
array(['VH: vertical transmit, horizontal receive', 'VV: vertical transmit, vertical receive'], dtype='<U41') - description(band)<U173'Terrain-corrected gamma naught ...
array(['Terrain-corrected gamma naught values of signal transmitted with vertical polarization and received with horizontal polarization with radiometric terrain correction applied.', 'Terrain-corrected gamma naught values of signal transmitted with vertical polarization and received with vertical polarization with radiometric terrain correction applied.'], dtype='<U173') - raster:bands()object{'nodata': -32768, 'data_type': ...
array({'nodata': -32768, 'data_type': 'float32', 'spatial_resolution': 10.0}, dtype=object) - epsg()int6432645
array(32645)
- vv(time, y, x, band)float64dask.array<chunksize=(1, 396, 290, 1), meta=np.ndarray>
Array Chunk Bytes 145.44 MiB 897.19 kiB Shape (83, 396, 290, 2) (1, 396, 290, 1) Count 42 Graph Layers 166 Chunks Type float64 numpy.ndarray - vh(time, y, x, band)float64dask.array<chunksize=(1, 396, 290, 1), meta=np.ndarray>
Array Chunk Bytes 145.44 MiB 897.19 kiB Shape (83, 396, 290, 2) (1, 396, 290, 1) Count 53 Graph Layers 166 Chunks Type float64 numpy.ndarray - ls(time, y, x, band)float64dask.array<chunksize=(1, 396, 290, 1), meta=np.ndarray>
Array Chunk Bytes 145.44 MiB 897.19 kiB Shape (83, 396, 290, 2) (1, 396, 290, 1) Count 57 Graph Layers 166 Chunks Type float64 numpy.ndarray
Great! Looks like its working
asf_subset
<xarray.Dataset>
Dimensions: (time: 83, y: 396, x: 290)
Coordinates: (12/20)
* time (time) datetime64[ns] 2021-06-02 ... 20...
granule_id (time) <U67 'S1A_IW_SLC__1SDV_20210602T...
* x (x) float64 6.194e+05 ... 6.281e+05
* y (y) float64 3.102e+06 ... 3.09e+06
spatial_ref int64 0
sensor (time) <U3 'S1A' 'S1A' ... 'S1A' 'S1A'
... ...
filtered (time) <U1 'n' 'n' 'n' 'n' ... 'n' 'n' 'n'
area (time) <U1 'e' 'e' 'e' 'e' ... 'e' 'e' 'e'
product_id (time) <U4 '1424' 'ABA0' ... '5BAD' 'BD36'
acq_hour (time) int64 12 12 0 12 0 ... 12 0 0 0 0
orbital_dir (time) <U4 'desc' 'desc' ... 'asc' 'asc'
data_take_id (time) <U6 '0480FD' '048318' ... '052C00'
Data variables:
vv (time, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
vh (time, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
ls (time, y, x) float64 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>- time: 83
- y: 396
- x: 290
- time(time)datetime64[ns]2021-06-02 ... 2022-05-21
array(['2021-06-02T00:00:00.000000000', '2021-06-07T00:00:00.000000000', '2021-06-10T00:00:00.000000000', '2021-06-14T00:00:00.000000000', '2021-06-19T00:00:00.000000000', '2021-06-22T00:00:00.000000000', '2021-06-26T00:00:00.000000000', '2021-07-01T00:00:00.000000000', '2021-07-04T00:00:00.000000000', '2021-07-08T00:00:00.000000000', '2021-07-16T00:00:00.000000000', '2021-07-20T00:00:00.000000000', '2021-07-25T00:00:00.000000000', '2021-07-28T00:00:00.000000000', '2021-08-01T00:00:00.000000000', '2021-08-06T00:00:00.000000000', '2021-08-09T00:00:00.000000000', '2021-08-13T00:00:00.000000000', '2021-08-18T00:00:00.000000000', '2021-08-21T00:00:00.000000000', '2021-08-25T00:00:00.000000000', '2021-09-02T00:00:00.000000000', '2021-09-06T00:00:00.000000000', '2021-09-11T00:00:00.000000000', '2021-09-14T00:00:00.000000000', '2021-09-18T00:00:00.000000000', '2021-09-23T00:00:00.000000000', '2021-09-26T00:00:00.000000000', '2021-09-30T00:00:00.000000000', '2021-10-08T00:00:00.000000000', '2021-10-12T00:00:00.000000000', '2021-10-17T00:00:00.000000000', '2021-10-20T00:00:00.000000000', '2021-10-24T00:00:00.000000000', '2021-10-29T00:00:00.000000000', '2021-11-01T00:00:00.000000000', '2021-11-05T00:00:00.000000000', '2021-11-10T00:00:00.000000000', '2021-11-13T00:00:00.000000000', '2021-11-17T00:00:00.000000000', '2021-11-22T00:00:00.000000000', '2021-11-25T00:00:00.000000000', '2021-11-29T00:00:00.000000000', '2021-12-04T00:00:00.000000000', '2021-12-07T00:00:00.000000000', '2021-12-11T00:00:00.000000000', '2021-12-16T00:00:00.000000000', '2021-12-19T00:00:00.000000000', '2021-12-23T00:00:00.000000000', '2021-12-28T00:00:00.000000000', '2021-12-31T00:00:00.000000000', '2022-01-04T00:00:00.000000000', '2022-01-09T00:00:00.000000000', '2022-01-12T00:00:00.000000000', '2022-01-16T00:00:00.000000000', '2022-01-21T00:00:00.000000000', '2022-01-24T00:00:00.000000000', '2022-01-28T00:00:00.000000000', '2022-02-02T00:00:00.000000000', '2022-02-14T00:00:00.000000000', '2022-02-17T00:00:00.000000000', '2022-02-21T00:00:00.000000000', '2022-02-26T00:00:00.000000000', '2022-03-01T00:00:00.000000000', '2022-03-05T00:00:00.000000000', '2022-03-10T00:00:00.000000000', '2022-03-13T00:00:00.000000000', '2022-03-17T00:00:00.000000000', '2022-03-22T00:00:00.000000000', '2022-03-25T00:00:00.000000000', '2022-03-29T00:00:00.000000000', '2022-04-03T00:00:00.000000000', '2022-04-06T00:00:00.000000000', '2022-04-10T00:00:00.000000000', '2022-04-15T00:00:00.000000000', '2022-04-18T00:00:00.000000000', '2022-04-22T00:00:00.000000000', '2022-04-27T00:00:00.000000000', '2022-04-30T00:00:00.000000000', '2022-05-04T00:00:00.000000000', '2022-05-09T00:00:00.000000000', '2022-05-16T00:00:00.000000000', '2022-05-21T00:00:00.000000000'], dtype='datetime64[ns]') - granule_id(time)<U67'S1A_IW_SLC__1SDV_20210602T12054...
- description :
- source granule ID for ASF-processed S1 RTC imagery, extracted from README files for each scene
array(['S1A_IW_SLC__1SDV_20210602T120543_20210602T120610_038161_0480FD_893F', 'S1A_IW_SLC__1SDV_20210607T121351_20210607T121418_038234_048318_9B64', 'S1A_IW_SLC__1SDV_20210610T000310_20210610T000337_038270_04841E_601A', 'S1A_IW_SLC__1SDV_20210614T120544_20210614T120611_038336_04862F_337B', 'S1A_IW_SLC__1SDV_20210619T121352_20210619T121419_038409_04884C_1DBA', 'S1A_IW_SLC__1SDV_20210622T000310_20210622T000337_038445_04895A_842E', 'S1A_IW_SLC__1SDV_20210626T120545_20210626T120612_038511_048B6C_9585', 'S1A_IW_SLC__1SDV_20210701T121352_20210701T121419_038584_048D87_F95C', 'S1A_IW_SLC__1SDV_20210704T000311_20210704T000338_038620_048E99_1BAA', 'S1A_IW_SLC__1SDV_20210708T120545_20210708T120612_038686_0490AD_DD9E', 'S1A_IW_SLC__1SDV_20210716T000312_20210716T000339_038795_0493DB_53CD', 'S1A_IW_SLC__1SDV_20210720T120546_20210720T120613_038861_0495EC_C9D4', 'S1A_IW_SLC__1SDV_20210725T121354_20210725T121421_038934_04980F_1651', 'S1A_IW_SLC__1SDV_20210728T000312_20210728T000339_038970_04991E_C669', 'S1A_IW_SLC__1SDV_20210801T120547_20210801T120614_039036_049B1F_9997', 'S1A_IW_SLC__1SDV_20210806T121354_20210806T121421_039109_049D70_B0A0', 'S1A_IW_SLC__1SDV_20210809T000313_20210809T000340_039145_049EAC_AA8E', 'S1A_IW_SLC__1SDV_20210813T120548_20210813T120614_039211_04A0FF_405F', 'S1A_IW_SLC__1SDV_20210818T121355_20210818T121422_039284_04A383_54D5', 'S1A_IW_SLC__1SDV_20210821T000314_20210821T000341_039320_04A4B9_0878', ... 'S1A_IW_SLC__1SDV_20220305T120545_20220305T120612_042186_0506F7_1601', 'S1A_IW_SLC__1SDV_20220310T121353_20220310T121420_042259_050962_068B', 'S1A_IW_SLC__1SDV_20220313T000311_20220313T000338_042295_050AA2_2909', 'S1A_IW_SLC__1SDV_20220317T120545_20220317T120612_042361_050CE5_4CCF', 'S1A_IW_SLC__1SDV_20220322T121353_20220322T121420_042434_050F59_A1B6', 'S1A_IW_SLC__1SDV_20220325T000312_20220325T000339_042470_051096_BA6F', 'S1A_IW_SLC__1SDV_20220329T120546_20220329T120613_042536_0512D7_425A', 'S1A_IW_SLC__1SDV_20220403T121353_20220403T121420_042609_05154A_3FE7', 'S1A_IW_SLC__1SDV_20220406T000312_20220406T000339_042645_051682_4B40', 'S1A_IW_SLC__1SDV_20220410T120546_20220410T120613_042711_0518C0_3349', 'S1A_IW_SLC__1SDV_20220415T121353_20220415T121420_042784_051B30_EB87', 'S1A_IW_SLC__1SDV_20220418T000312_20220418T000339_042820_051C63_CAB2', 'S1A_IW_SLC__1SDV_20220422T120547_20220422T120614_042886_051E96_41EE', 'S1A_IW_SLC__1SDV_20220427T121354_20220427T121421_042959_0520EE_A42D', 'S1A_IW_SLC__1SDV_20220430T000313_20220430T000340_042995_05221E_46A1', 'S1A_IW_SLC__1SDV_20220504T120547_20220504T120614_043061_05245F_6F62', 'S1A_IW_SLC__1SDV_20220509T121355_20220509T121422_043134_0526C4_092E', 'S1A_IW_SLC__1SDV_20220516T120548_20220516T120615_043236_0529DD_42D2', 'S1A_IW_SLC__1SDV_20220521T121356_20220521T121423_043309_052C00_FE88'], dtype='<U67') - x(x)float646.194e+05 6.195e+05 ... 6.281e+05
- axis :
- X
- long_name :
- x coordinate of projection
- standard_name :
- projection_x_coordinate
- units :
- metre
array([619425., 619455., 619485., ..., 628035., 628065., 628095.])
- y(y)float643.102e+06 3.102e+06 ... 3.09e+06
- axis :
- Y
- long_name :
- y coordinate of projection
- standard_name :
- projection_y_coordinate
- units :
- metre
array([3101655., 3101625., 3101595., ..., 3089865., 3089835., 3089805.])
- spatial_ref()int640
- crs_wkt :
- PROJCS["WGS 84 / UTM zone 45N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",87],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32645"]]
- semi_major_axis :
- 6378137.0
- semi_minor_axis :
- 6356752.314245179
- inverse_flattening :
- 298.257223563
- reference_ellipsoid_name :
- WGS 84
- longitude_of_prime_meridian :
- 0.0
- prime_meridian_name :
- Greenwich
- geographic_crs_name :
- WGS 84
- horizontal_datum_name :
- World Geodetic System 1984
- projected_crs_name :
- WGS 84 / UTM zone 45N
- grid_mapping_name :
- transverse_mercator
- latitude_of_projection_origin :
- 0.0
- longitude_of_central_meridian :
- 87.0
- false_easting :
- 500000.0
- false_northing :
- 0.0
- scale_factor_at_central_meridian :
- 0.9996
- spatial_ref :
- PROJCS["WGS 84 / UTM zone 45N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",87],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32645"]]
- GeoTransform :
- 619410.0 30.0 0.0 3101670.0 0.0 -30.0
array(0)
- sensor(time)<U3'S1A' 'S1A' 'S1A' ... 'S1A' 'S1A'
array(['S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A'], dtype='<U3') - beam_mode(time)<U2'IW' 'IW' 'IW' ... 'IW' 'IW' 'IW'
array(['IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW'], dtype='<U2') - acquisition_time(time)datetime64[ns]2022-10-21T12:14:14 ... 2022-10-...
array(['2022-10-21T12:14:14.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:14:20.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T12:05:43.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:05:43.000000000', '2022-10-21T00:03:14.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:52.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T12:13:50.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T12:14:17.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T00:03:10.000000000', '2022-10-21T12:05:45.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:05:50.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:52.000000000', '2022-10-21T00:03:08.000000000', ... '2022-10-21T12:05:42.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:07.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:50.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T00:03:14.000000000', '2022-10-21T12:05:44.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:13:49.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:45.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T00:03:16.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T12:13:51.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T00:03:09.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T00:03:12.000000000'], dtype='datetime64[ns]') - polarisation_type(time)<U2'DV' 'DV' 'DV' ... 'DV' 'DV' 'DV'
array(['DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV'], dtype='<U2') - orbit_type(time)<U1'P' 'P' 'P' 'P' ... 'P' 'P' 'P' 'P'
array(['P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P'], dtype='<U1') - terrain_correction_pixel_spacing(time)<U5'RTC30' 'RTC30' ... 'RTC30' 'RTC30'
array(['RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30'], dtype='<U5') - output_format(time)<U1'g' 'g' 'g' 'g' ... 'g' 'g' 'g' 'g'
array(['g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g'], dtype='<U1') - output_type(time)<U1'p' 'p' 'p' 'p' ... 'p' 'p' 'p' 'p'
array(['p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p'], dtype='<U1') - masked(time)<U1'u' 'u' 'u' 'u' ... 'u' 'u' 'u' 'u'
array(['u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u'], dtype='<U1') - filtered(time)<U1'n' 'n' 'n' 'n' ... 'n' 'n' 'n' 'n'
array(['n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n'], dtype='<U1') - area(time)<U1'e' 'e' 'e' 'e' ... 'e' 'e' 'e' 'e'
array(['e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e'], dtype='<U1') - product_id(time)<U4'1424' 'ABA0' ... '5BAD' 'BD36'
array(['1424', 'ABA0', 'E61E', 'E5B6', '5569', '71C9', 'B942', 'DCE1', '6581', '1DFD', '01B2', '9849', 'CFFC', '57F2', 'D4E6', 'B7B0', '3770', '52BB', '0579', 'AA68', '24B8', '5FF4', '5F4E', '199D', '0031', 'D082', 'E74B', 'F7ED', '65E0', '528F', '51E7', 'F5AA', 'AC38', '772E', 'F0A6', '066E', '5844', '8CD8', '6340', 'D35A', '056C', '6E6F', '0CAD', '6270', '8A4F', 'BEAB', '54B1', '8F7B', 'F9A9', 'AA59', '57E1', '81EB', '250E', '3C39', '3A2A', '9331', 'C14C', 'F1DE', '106F', 'FD00', '5D96', '971C', 'F573', 'C890', '68EA', '83D7', 'E113', 'D846', 'F4B3', 'BB26', '485C', '6683', '6153', '832B', '7418', '378D', '33F5', '1139', 'B661', '9A2F', '6943', '5BAD', 'BD36'], dtype='<U4') - acq_hour(time)int6412 12 0 12 0 12 0 ... 12 12 0 0 0 0
array([12, 12, 0, 12, 0, 12, 0, 12, 0, 12, 0, 12, 12, 12, 0, 0, 12, 0, 12, 12, 12, 12, 0, 0, 12, 0, 12, 12, 12, 12, 12, 12, 0, 12, 12, 12, 12, 0, 12, 0, 12, 12, 12, 12, 12, 12, 0, 12, 12, 0, 12, 12, 12, 12, 12, 0, 12, 12, 0, 12, 12, 12, 12, 12, 0, 12, 12, 0, 0, 0, 12, 12, 12, 12, 12, 12, 12, 12, 12, 0, 0, 0, 0]) - orbital_dir(time)<U4'desc' 'desc' 'asc' ... 'asc' 'asc'
array(['desc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'asc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'asc', 'asc'], dtype='<U4') - data_take_id(time)<U6'0480FD' '048318' ... '052C00'
- ID of data take from SAR acquisition :
- ID data take
array(['0480FD', '048318', '04841E', '04862F', '04884C', '04895A', '048B6C', '048D87', '048E99', '0490AD', '0493DB', '0495EC', '04980F', '04991E', '049B1F', '049D70', '049EAC', '04A0FF', '04A383', '04A4B9', '04A6FC', '04AAB3', '04AD0D', '04AF85', '04B0C4', '04B2FB', '04B566', '04B6AB', '04B8FF', '04BCB8', '04BF12', '04C195', '04C2D6', '04C52E', '04C7A8', '04C8E9', '04CB46', '04CDC8', '04CEFF', '04D14F', '04D3CC', '04D50E', '04D761', '04D9E8', '04DB21', '04DD71', '04DFDD', '04E110', '04E340', '04E5AB', '04E6D7', '04E931', '04EB9B', '04ECCD', '04EEE8', '04F156', '04F294', '04F4D6', '04F754', '04FD6F', '04FEB2', '050108', '050376', '0504AF', '0506F7', '050962', '050AA2', '050CE5', '050F59', '051096', '0512D7', '05154A', '051682', '0518C0', '051B30', '051C63', '051E96', '0520EE', '05221E', '05245F', '0526C4', '0529DD', '052C00'], dtype='<U6')
- vv(time, y, x)float32dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
- scale_factor :
- 1.0
- add_offset :
- 0.0
- _FillValue :
- 0.0
Array Chunk Bytes 36.36 MiB 448.59 kiB Shape (83, 396, 290) (1, 396, 290) Count 19 Graph Layers 83 Chunks Type float32 numpy.ndarray - vh(time, y, x)float32dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
- scale_factor :
- 1.0
- add_offset :
- 0.0
- _FillValue :
- 0.0
Array Chunk Bytes 36.36 MiB 448.59 kiB Shape (83, 396, 290) (1, 396, 290) Count 30 Graph Layers 83 Chunks Type float32 numpy.ndarray - ls(time, y, x)float64dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
- scale_factor :
- 1.0
- add_offset :
- 0.0
- _FillValue :
- 0
Array Chunk Bytes 72.72 MiB 897.19 kiB Shape (83, 396, 290) (1, 396, 290) Count 34 Graph Layers 83 Chunks Type float64 numpy.ndarray
pc_mask.sel(band='vv').mean(dim=['x','y'])
<xarray.Dataset>
Dimensions: (time: 83)
Coordinates: (12/53)
* time (time) datetime64[ns] 2021-06-02 ....
spatial_ref int64 0
sensor (time) <U3 'S1A' 'S1A' ... 'S1A'
beam_mode (time) <U2 'IW' 'IW' ... 'IW' 'IW'
acquisition_time (time) datetime64[ns] 2022-10-21T1...
polarisation_type (time) <U2 'DV' 'DV' ... 'DV' 'DV'
... ...
s1:total_slices (time) <U2 '21' '20' ... '21' '20'
s1:processing_level <U1 '1'
title <U41 'VV: vertical transmit, verti...
description <U173 'Terrain-corrected gamma nau...
raster:bands object {'nodata': -32768, 'data_ty...
epsg int64 32645
Data variables:
vv (time) float64 dask.array<chunksize=(1,), meta=np.ndarray>
vh (time) float64 dask.array<chunksize=(1,), meta=np.ndarray>
ls (time) float64 dask.array<chunksize=(1,), meta=np.ndarray>- time: 83
- time(time)datetime64[ns]2021-06-02 ... 2022-05-21
array(['2021-06-02T00:00:00.000000000', '2021-06-07T00:00:00.000000000', '2021-06-10T00:00:00.000000000', '2021-06-14T00:00:00.000000000', '2021-06-19T00:00:00.000000000', '2021-06-22T00:00:00.000000000', '2021-06-26T00:00:00.000000000', '2021-07-01T00:00:00.000000000', '2021-07-04T00:00:00.000000000', '2021-07-08T00:00:00.000000000', '2021-07-16T00:00:00.000000000', '2021-07-20T00:00:00.000000000', '2021-07-25T00:00:00.000000000', '2021-07-28T00:00:00.000000000', '2021-08-01T00:00:00.000000000', '2021-08-06T00:00:00.000000000', '2021-08-09T00:00:00.000000000', '2021-08-13T00:00:00.000000000', '2021-08-18T00:00:00.000000000', '2021-08-21T00:00:00.000000000', '2021-08-25T00:00:00.000000000', '2021-09-02T00:00:00.000000000', '2021-09-06T00:00:00.000000000', '2021-09-11T00:00:00.000000000', '2021-09-14T00:00:00.000000000', '2021-09-18T00:00:00.000000000', '2021-09-23T00:00:00.000000000', '2021-09-26T00:00:00.000000000', '2021-09-30T00:00:00.000000000', '2021-10-08T00:00:00.000000000', '2021-10-12T00:00:00.000000000', '2021-10-17T00:00:00.000000000', '2021-10-20T00:00:00.000000000', '2021-10-24T00:00:00.000000000', '2021-10-29T00:00:00.000000000', '2021-11-01T00:00:00.000000000', '2021-11-05T00:00:00.000000000', '2021-11-10T00:00:00.000000000', '2021-11-13T00:00:00.000000000', '2021-11-17T00:00:00.000000000', '2021-11-22T00:00:00.000000000', '2021-11-25T00:00:00.000000000', '2021-11-29T00:00:00.000000000', '2021-12-04T00:00:00.000000000', '2021-12-07T00:00:00.000000000', '2021-12-11T00:00:00.000000000', '2021-12-16T00:00:00.000000000', '2021-12-19T00:00:00.000000000', '2021-12-23T00:00:00.000000000', '2021-12-28T00:00:00.000000000', '2021-12-31T00:00:00.000000000', '2022-01-04T00:00:00.000000000', '2022-01-09T00:00:00.000000000', '2022-01-12T00:00:00.000000000', '2022-01-16T00:00:00.000000000', '2022-01-21T00:00:00.000000000', '2022-01-24T00:00:00.000000000', '2022-01-28T00:00:00.000000000', '2022-02-02T00:00:00.000000000', '2022-02-14T00:00:00.000000000', '2022-02-17T00:00:00.000000000', '2022-02-21T00:00:00.000000000', '2022-02-26T00:00:00.000000000', '2022-03-01T00:00:00.000000000', '2022-03-05T00:00:00.000000000', '2022-03-10T00:00:00.000000000', '2022-03-13T00:00:00.000000000', '2022-03-17T00:00:00.000000000', '2022-03-22T00:00:00.000000000', '2022-03-25T00:00:00.000000000', '2022-03-29T00:00:00.000000000', '2022-04-03T00:00:00.000000000', '2022-04-06T00:00:00.000000000', '2022-04-10T00:00:00.000000000', '2022-04-15T00:00:00.000000000', '2022-04-18T00:00:00.000000000', '2022-04-22T00:00:00.000000000', '2022-04-27T00:00:00.000000000', '2022-04-30T00:00:00.000000000', '2022-05-04T00:00:00.000000000', '2022-05-09T00:00:00.000000000', '2022-05-16T00:00:00.000000000', '2022-05-21T00:00:00.000000000'], dtype='datetime64[ns]') - spatial_ref()int640
array(0)
- sensor(time)<U3'S1A' 'S1A' 'S1A' ... 'S1A' 'S1A'
array(['S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A'], dtype='<U3') - beam_mode(time)<U2'IW' 'IW' 'IW' ... 'IW' 'IW' 'IW'
array(['IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW'], dtype='<U2') - acquisition_time(time)datetime64[ns]2022-10-21T12:14:14 ... 2022-10-...
array(['2022-10-21T12:14:14.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:14:20.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T12:05:43.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:05:43.000000000', '2022-10-21T00:03:14.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:52.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T12:13:50.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T12:14:17.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T00:03:10.000000000', '2022-10-21T12:05:45.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:05:50.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:52.000000000', '2022-10-21T00:03:08.000000000', ... '2022-10-21T12:05:42.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:07.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:50.000000000', '2022-10-21T12:13:53.000000000', '2022-10-21T12:13:56.000000000', '2022-10-21T00:03:14.000000000', '2022-10-21T12:05:44.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:05:46.000000000', '2022-10-21T12:13:49.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:45.000000000', '2022-10-21T00:03:15.000000000', '2022-10-21T12:13:57.000000000', '2022-10-21T12:14:18.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T00:03:12.000000000', '2022-10-21T00:03:16.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T12:05:49.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T12:05:48.000000000', '2022-10-21T12:05:47.000000000', '2022-10-21T12:13:55.000000000', '2022-10-21T12:13:51.000000000', '2022-10-21T12:13:54.000000000', '2022-10-21T00:03:09.000000000', '2022-10-21T00:03:11.000000000', '2022-10-21T00:03:13.000000000', '2022-10-21T00:03:12.000000000'], dtype='datetime64[ns]') - polarisation_type(time)<U2'DV' 'DV' 'DV' ... 'DV' 'DV' 'DV'
array(['DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV'], dtype='<U2') - orbit_type(time)<U1'P' 'P' 'P' 'P' ... 'P' 'P' 'P' 'P'
array(['P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P'], dtype='<U1') - terrain_correction_pixel_spacing(time)<U5'RTC30' 'RTC30' ... 'RTC30' 'RTC30'
array(['RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30'], dtype='<U5') - output_format(time)<U1'g' 'g' 'g' 'g' ... 'g' 'g' 'g' 'g'
array(['g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g'], dtype='<U1') - output_type(time)<U1'p' 'p' 'p' 'p' ... 'p' 'p' 'p' 'p'
array(['p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p'], dtype='<U1') - masked(time)<U1'u' 'u' 'u' 'u' ... 'u' 'u' 'u' 'u'
array(['u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u'], dtype='<U1') - filtered(time)<U1'n' 'n' 'n' 'n' ... 'n' 'n' 'n' 'n'
array(['n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n'], dtype='<U1') - area(time)<U1'e' 'e' 'e' 'e' ... 'e' 'e' 'e' 'e'
array(['e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e'], dtype='<U1') - product_id(time)<U4'1424' 'ABA0' ... '5BAD' 'BD36'
array(['1424', 'ABA0', 'E61E', 'E5B6', '5569', '71C9', 'B942', 'DCE1', '6581', '1DFD', '01B2', '9849', 'CFFC', '57F2', 'D4E6', 'B7B0', '3770', '52BB', '0579', 'AA68', '24B8', '5FF4', '5F4E', '199D', '0031', 'D082', 'E74B', 'F7ED', '65E0', '528F', '51E7', 'F5AA', 'AC38', '772E', 'F0A6', '066E', '5844', '8CD8', '6340', 'D35A', '056C', '6E6F', '0CAD', '6270', '8A4F', 'BEAB', '54B1', '8F7B', 'F9A9', 'AA59', '57E1', '81EB', '250E', '3C39', '3A2A', '9331', 'C14C', 'F1DE', '106F', 'FD00', '5D96', '971C', 'F573', 'C890', '68EA', '83D7', 'E113', 'D846', 'F4B3', 'BB26', '485C', '6683', '6153', '832B', '7418', '378D', '33F5', '1139', 'B661', '9A2F', '6943', '5BAD', 'BD36'], dtype='<U4') - acq_hour(time)int6412 12 0 12 0 12 0 ... 12 12 0 0 0 0
array([12, 12, 0, 12, 0, 12, 0, 12, 0, 12, 0, 12, 12, 12, 0, 0, 12, 0, 12, 12, 12, 12, 0, 0, 12, 0, 12, 12, 12, 12, 12, 12, 0, 12, 12, 12, 12, 0, 12, 0, 12, 12, 12, 12, 12, 12, 0, 12, 12, 0, 12, 12, 12, 12, 12, 0, 12, 12, 0, 12, 12, 12, 12, 12, 0, 12, 12, 0, 0, 0, 12, 12, 12, 12, 12, 12, 12, 12, 12, 0, 0, 0, 0]) - orbital_dir(time)<U4'desc' 'desc' 'asc' ... 'asc' 'asc'
array(['desc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'asc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'asc', 'asc'], dtype='<U4') - data_take_id(time)<U6'0480FD' '048318' ... '052C00'
array(['0480FD', '048318', '04841E', '04862F', '04884C', '04895A', '048B6C', '048D87', '048E99', '0490AD', '0493DB', '0495EC', '04980F', '04991E', '049B1F', '049D70', '049EAC', '04A0FF', '04A383', '04A4B9', '04A6FC', '04AAB3', '04AD0D', '04AF85', '04B0C4', '04B2FB', '04B566', '04B6AB', '04B8FF', '04BCB8', '04BF12', '04C195', '04C2D6', '04C52E', '04C7A8', '04C8E9', '04CB46', '04CDC8', '04CEFF', '04D14F', '04D3CC', '04D50E', '04D761', '04D9E8', '04DB21', '04DD71', '04DFDD', '04E110', '04E340', '04E5AB', '04E6D7', '04E931', '04EB9B', '04ECCD', '04EEE8', '04F156', '04F294', '04F4D6', '04F754', '04FD6F', '04FEB2', '050108', '050376', '0504AF', '0506F7', '050962', '050AA2', '050CE5', '050F59', '051096', '0512D7', '05154A', '051682', '0518C0', '051B30', '051C63', '051E96', '0520EE', '05221E', '05245F', '0526C4', '0529DD', '052C00'], dtype='<U6') - id(time)<U66'S1A_IW_GRDH_1SDV_20210602T12054...
array(['S1A_IW_GRDH_1SDV_20210602T120544_20210602T120609_038161_0480FD_rtc', 'S1A_IW_GRDH_1SDV_20210607T121352_20210607T121417_038234_048318_rtc', 'S1A_IW_GRDH_1SDV_20210610T000311_20210610T000336_038270_04841E_rtc', 'S1A_IW_GRDH_1SDV_20210614T120545_20210614T120610_038336_04862F_rtc', 'S1A_IW_GRDH_1SDV_20210619T121353_20210619T121418_038409_04884C_rtc', 'S1A_IW_GRDH_1SDV_20210622T000312_20210622T000337_038445_04895A_rtc', 'S1A_IW_GRDH_1SDV_20210626T120546_20210626T120611_038511_048B6C_rtc', 'S1A_IW_GRDH_1SDV_20210701T121354_20210701T121419_038584_048D87_rtc', 'S1A_IW_GRDH_1SDV_20210704T000312_20210704T000337_038620_048E99_rtc', 'S1A_IW_GRDH_1SDV_20210708T120547_20210708T120612_038686_0490AD_rtc', 'S1A_IW_GRDH_1SDV_20210716T000313_20210716T000338_038795_0493DB_rtc', 'S1A_IW_GRDH_1SDV_20210720T120547_20210720T120612_038861_0495EC_rtc', 'S1A_IW_GRDH_1SDV_20210725T121355_20210725T121420_038934_04980F_rtc', 'S1A_IW_GRDH_1SDV_20210728T000314_20210728T000339_038970_04991E_rtc', 'S1A_IW_GRDH_1SDV_20210801T120548_20210801T120613_039036_049B1F_rtc', 'S1A_IW_GRDH_1SDV_20210806T121356_20210806T121421_039109_049D70_rtc', 'S1A_IW_GRDH_1SDV_20210809T000314_20210809T000339_039145_049EAC_rtc', 'S1A_IW_GRDH_1SDV_20210813T120549_20210813T120614_039211_04A0FF_rtc', 'S1A_IW_GRDH_1SDV_20210818T121356_20210818T121421_039284_04A383_rtc', 'S1A_IW_GRDH_1SDV_20210821T000315_20210821T000340_039320_04A4B9_rtc', ... 'S1A_IW_GRDH_1SDV_20220305T120547_20220305T120612_042186_0506F7_rtc', 'S1A_IW_GRDH_1SDV_20220310T121354_20220310T121419_042259_050962_rtc', 'S1A_IW_GRDH_1SDV_20220313T000313_20220313T000338_042295_050AA2_rtc', 'S1A_IW_GRDH_1SDV_20220317T120547_20220317T120612_042361_050CE5_rtc', 'S1A_IW_GRDH_1SDV_20220322T121354_20220322T121419_042434_050F59_rtc', 'S1A_IW_GRDH_1SDV_20220325T000313_20220325T000338_042470_051096_rtc', 'S1A_IW_GRDH_1SDV_20220329T120547_20220329T120612_042536_0512D7_rtc', 'S1A_IW_GRDH_1SDV_20220403T121355_20220403T121420_042609_05154A_rtc', 'S1A_IW_GRDH_1SDV_20220406T000313_20220406T000338_042645_051682_rtc', 'S1A_IW_GRDH_1SDV_20220410T120547_20220410T120612_042711_0518C0_rtc', 'S1A_IW_GRDH_1SDV_20220415T121355_20220415T121420_042784_051B30_rtc', 'S1A_IW_GRDH_1SDV_20220418T000314_20220418T000339_042820_051C63_rtc', 'S1A_IW_GRDH_1SDV_20220422T120548_20220422T120613_042886_051E96_rtc', 'S1A_IW_GRDH_1SDV_20220427T121355_20220427T121420_042959_0520EE_rtc', 'S1A_IW_GRDH_1SDV_20220430T000314_20220430T000339_042995_05221E_rtc', 'S1A_IW_GRDH_1SDV_20220504T120548_20220504T120613_043061_05245F_rtc', 'S1A_IW_GRDH_1SDV_20220509T121356_20220509T121421_043134_0526C4_rtc', 'S1A_IW_GRDH_1SDV_20220516T120549_20220516T120614_043236_0529DD_rtc', 'S1A_IW_GRDH_1SDV_20220521T121357_20220521T121422_043309_052C00_rtc'], dtype='<U66') - band()<U2'vv'
array('vv', dtype='<U2') - sar:looks_azimuth()int641
array(1)
- sat:relative_orbit(time)int64114 12 48 114 12 ... 114 12 114 12
array([114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 48, 114, 12, 114, 12]) - s1:instrument_configuration_ID(time)<U1'6' '6' '6' '6' ... '7' '7' '7' '7'
array(['6', '6', '6', '6', '6', '6', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7', '7'], dtype='<U1') - s1:orbit_source(time)<U8'DOWNLINK' 'DOWNLINK' ... 'RESORB'
array(['DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'DOWNLINK', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'PREORB', 'PREORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB', 'RESORB'], dtype='<U8') - end_datetime(time)<U32'2021-06-02 12:06:09.940813+00:0...
array(['2021-06-02 12:06:09.940813+00:00', '2021-06-07 12:14:17.557268+00:00', '2021-06-10 00:03:36.369821+00:00', '2021-06-14 12:06:10.872859+00:00', '2021-06-19 12:14:18.365410+00:00', '2021-06-22 00:03:37.093898+00:00', '2021-06-26 12:06:11.482149+00:00', '2021-07-01 12:14:19.071405+00:00', '2021-07-04 00:03:37.847082+00:00', '2021-07-08 12:06:12.112559+00:00', '2021-07-16 00:03:38.592472+00:00', '2021-07-20 12:06:12.927516+00:00', '2021-07-25 12:14:20.313830+00:00', '2021-07-28 00:03:39.166376+00:00', '2021-08-01 12:06:13.649807+00:00', '2021-08-06 12:14:21.197519+00:00', '2021-08-09 00:03:39.953924+00:00', '2021-08-13 12:06:14.213502+00:00', '2021-08-18 12:14:21.725577+00:00', '2021-08-21 00:03:40.559065+00:00', ... '2022-03-01 00:03:38.089393+00:00', '2022-03-05 12:06:12.176227+00:00', '2022-03-10 12:14:19.493699+00:00', '2022-03-13 00:03:38.244245+00:00', '2022-03-17 12:06:12.205265+00:00', '2022-03-22 12:14:19.861261+00:00', '2022-03-25 00:03:38.638828+00:00', '2022-03-29 12:06:12.640119+00:00', '2022-04-03 12:14:20.084951+00:00', '2022-04-06 00:03:38.681256+00:00', '2022-04-10 12:06:12.897487+00:00', '2022-04-15 12:14:20.210307+00:00', '2022-04-18 00:03:39.102936+00:00', '2022-04-22 12:06:13.551041+00:00', '2022-04-27 12:14:20.924939+00:00', '2022-04-30 00:03:39.701264+00:00', '2022-05-04 12:06:13.622093+00:00', '2022-05-09 12:14:21.479341+00:00', '2022-05-16 12:06:14.814536+00:00', '2022-05-21 12:14:22.285304+00:00'], dtype='<U32') - sat:orbit_state(time)<U10'ascending' ... 'ascending'
array(['ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'descending', 'ascending', 'ascending', 'ascending', 'ascending'], dtype='<U10') - platform()<U11'SENTINEL-1A'
array('SENTINEL-1A', dtype='<U11') - s1:slice_number(time)<U2'6' '6' '12' '6' ... '6' '6' '6'
array(['6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '12', '6', '6', '6', '6'], dtype='<U2') - start_datetime(time)<U32'2021-06-02 12:05:44.941334+00:0...
array(['2021-06-02 12:05:44.941334+00:00', '2021-06-07 12:13:52.559418+00:00', '2021-06-10 00:03:11.371078+00:00', '2021-06-14 12:05:45.874802+00:00', '2021-06-19 12:13:53.365914+00:00', '2021-06-22 00:03:12.095344+00:00', '2021-06-26 12:05:46.482738+00:00', '2021-07-01 12:13:54.073541+00:00', '2021-07-04 00:03:12.848412+00:00', '2021-07-08 12:05:47.114550+00:00', '2021-07-16 00:03:13.593918+00:00', '2021-07-20 12:05:47.929430+00:00', '2021-07-25 12:13:55.316007+00:00', '2021-07-28 00:03:14.167671+00:00', '2021-08-01 12:05:48.651907+00:00', '2021-08-06 12:13:56.199672+00:00', '2021-08-09 00:03:14.955252+00:00', '2021-08-13 12:05:49.214084+00:00', '2021-08-18 12:13:56.727697+00:00', '2021-08-21 00:03:15.560368+00:00', ... '2022-03-01 00:03:13.090580+00:00', '2022-03-05 12:05:47.177919+00:00', '2022-03-10 12:13:54.495512+00:00', '2022-03-13 00:03:13.245549+00:00', '2022-03-17 12:05:47.206984+00:00', '2022-03-22 12:13:54.863098+00:00', '2022-03-25 00:03:13.640217+00:00', '2022-03-29 12:05:47.640405+00:00', '2022-04-03 12:13:55.086898+00:00', '2022-04-06 00:03:13.682743+00:00', '2022-04-10 12:05:47.899396+00:00', '2022-04-15 12:13:55.212427+00:00', '2022-04-18 00:03:14.104239+00:00', '2022-04-22 12:05:48.551577+00:00', '2022-04-27 12:13:55.927061+00:00', '2022-04-30 00:03:14.702542+00:00', '2022-05-04 12:05:48.622638+00:00', '2022-05-09 12:13:56.481455+00:00', '2022-05-16 12:05:49.815129+00:00', '2022-05-21 12:13:57.287447+00:00'], dtype='<U32') - sar:resolution_azimuth()int6422
array(22)
- s1:resolution()<U4'high'
array('high', dtype='<U4') - sar:frequency_band()<U1'C'
array('C', dtype='<U1') - sar:center_frequency()float645.405
array(5.405)
- sar:pixel_spacing_azimuth()int6410
array(10)
- sat:platform_international_designator()<U9'2014-016A'
array('2014-016A', dtype='<U9') - constellation()<U10'Sentinel-1'
array('Sentinel-1', dtype='<U10') - sar:polarizations()object{'VV', 'VH'}
array({'VV', 'VH'}, dtype=object) - proj:epsg()int6432645
array(32645)
- sar:pixel_spacing_range()int6410
array(10)
- sar:looks_range()int645
array(5)
- sar:resolution_range()int6420
array(20)
- sat:absolute_orbit(time)int6438161 38234 38270 ... 43236 43309
array([38161, 38234, 38270, 38336, 38409, 38445, 38511, 38584, 38620, 38686, 38795, 38861, 38934, 38970, 39036, 39109, 39145, 39211, 39284, 39320, 39386, 39495, 39561, 39634, 39670, 39736, 39809, 39845, 39911, 40020, 40086, 40159, 40195, 40261, 40334, 40370, 40436, 40509, 40545, 40611, 40684, 40720, 40786, 40859, 40895, 40961, 41034, 41070, 41136, 41209, 41245, 41311, 41384, 41420, 41486, 41559, 41595, 41661, 41734, 41909, 41945, 42011, 42084, 42120, 42186, 42259, 42295, 42361, 42434, 42470, 42536, 42609, 42645, 42711, 42784, 42820, 42886, 42959, 42995, 43061, 43134, 43236, 43309]) - sar:looks_equivalent_number()float644.4
array(4.4)
- sar:observation_direction()<U5'right'
array('right', dtype='<U5') - sar:product_type()<U3'GRD'
array('GRD', dtype='<U3') - s1:product_timeliness()<U8'Fast-24h'
array('Fast-24h', dtype='<U8') - sar:instrument_mode()<U2'IW'
array('IW', dtype='<U2') - s1:datatake_id(time)<U6'295165' '295704' ... '338944'
array(['295165', '295704', '295966', '296495', '297036', '297306', '297836', '298375', '298649', '299181', '299995', '300524', '301071', '301342', '301855', '302448', '302764', '303359', '304003', '304313', '304892', '305843', '306445', '307077', '307396', '307963', '308582', '308907', '309503', '310456', '311058', '311701', '312022', '312622', '313256', '313577', '314182', '314824', '315135', '315727', '316364', '316686', '317281', '317928', '318241', '318833', '319453', '319760', '320320', '320939', '321239', '321841', '322459', '322765', '323304', '323926', '324244', '324822', '325460', '327023', '327346', '327944', '328566', '328879', '329463', '330082', '330402', '330981', '331609', '331926', '332503', '333130', '333442', '334016', '334640', '334947', '335510', '336110', '336414', '336991', '337604', '338397', '338944'], dtype='<U6') - s1:total_slices(time)<U2'21' '20' '17' ... '20' '21' '20'
array(['21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '17', '21', '20', '21', '20'], dtype='<U2') - s1:processing_level()<U1'1'
array('1', dtype='<U1') - title()<U41'VV: vertical transmit, vertical...
array('VV: vertical transmit, vertical receive', dtype='<U41') - description()<U173'Terrain-corrected gamma naught ...
array('Terrain-corrected gamma naught values of signal transmitted with vertical polarization and received with vertical polarization with radiometric terrain correction applied.', dtype='<U173') - raster:bands()object{'nodata': -32768, 'data_type': ...
array({'nodata': -32768, 'data_type': 'float32', 'spatial_resolution': 10.0}, dtype=object) - epsg()int6432645
array(32645)
- vv(time)float64dask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 664 B 8 B Shape (83,) (1,) Count 45 Graph Layers 83 Chunks Type float64 numpy.ndarray - vh(time)float64dask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 664 B 8 B Shape (83,) (1,) Count 56 Graph Layers 83 Chunks Type float64 numpy.ndarray - ls(time)float64dask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 664 B 8 B Shape (83,) (1,) Count 60 Graph Layers 83 Chunks Type float64 numpy.ndarray
fig, ax = plt.subplots(figsize=(12,8))
power_to_db(asf_subset.vv.mean(dim=['x','y'])).plot(ax=ax, color='blue', linestyle='None', marker='o', markersize=5, label = 'ASF');
power_to_db(pc_mask.sel(band='vv').mean(dim=['x','y'], skipna=True).vv).plot(x = 'time',ax=ax, color='red', linestyle='None', marker='o', markersize=5, label='PC');
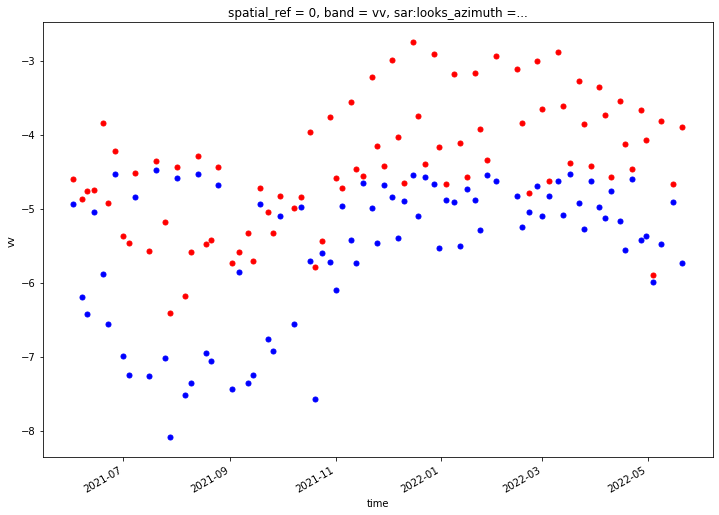
Break up by asc, desc pass#
Use xr.where() to subset datasets by orbital pass:
asf_subset
<xarray.Dataset>
Dimensions: (acq_date: 90, y: 396, x: 290)
Coordinates: (12/20)
* acq_date (acq_date) datetime64[ns] 2021-06-02 .....
granule_id (acq_date) <U67 'S1A_IW_SLC__1SDV_20210...
* x (x) float64 6.194e+05 ... 6.281e+05
* y (y) float64 3.102e+06 ... 3.09e+06
spatial_ref int64 0
sensor (acq_date) <U3 'S1A' 'S1A' ... 'S1A' 'S1A'
... ...
filtered (acq_date) <U1 'n' 'n' 'n' ... 'n' 'n' 'n'
area (acq_date) <U1 'e' 'e' 'e' ... 'e' 'e' 'e'
product_id (acq_date) <U4 '1424' 'ABA0' ... 'BD36'
acq_hour (acq_date) int64 12 12 0 12 ... 0 12 12 0
orbital_dir (acq_date) <U4 'desc' 'desc' ... 'asc'
data_take_id (acq_date) <U6 '0480FD' ... '052C00'
Data variables:
vv (acq_date, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
vh (acq_date, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
ls (acq_date, y, x) float64 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>- acq_date: 90
- y: 396
- x: 290
- acq_date(acq_date)datetime64[ns]2021-06-02 ... 2022-05-21
array(['2021-06-02T00:00:00.000000000', '2021-06-07T00:00:00.000000000', '2021-06-10T00:00:00.000000000', '2021-06-14T00:00:00.000000000', '2021-06-19T00:00:00.000000000', '2021-06-22T00:00:00.000000000', '2021-06-26T00:00:00.000000000', '2021-07-01T00:00:00.000000000', '2021-07-01T00:00:00.000000000', '2021-07-04T00:00:00.000000000', '2021-07-08T00:00:00.000000000', '2021-07-13T00:00:00.000000000', '2021-07-13T00:00:00.000000000', '2021-07-13T00:00:00.000000000', '2021-07-16T00:00:00.000000000', '2021-07-20T00:00:00.000000000', '2021-07-25T00:00:00.000000000', '2021-07-28T00:00:00.000000000', '2021-08-01T00:00:00.000000000', '2021-08-06T00:00:00.000000000', '2021-08-09T00:00:00.000000000', '2021-08-13T00:00:00.000000000', '2021-08-18T00:00:00.000000000', '2021-08-21T00:00:00.000000000', '2021-08-25T00:00:00.000000000', '2021-09-02T00:00:00.000000000', '2021-09-06T00:00:00.000000000', '2021-09-11T00:00:00.000000000', '2021-09-14T00:00:00.000000000', '2021-09-18T00:00:00.000000000', '2021-09-23T00:00:00.000000000', '2021-09-26T00:00:00.000000000', '2021-09-30T00:00:00.000000000', '2021-10-08T00:00:00.000000000', '2021-10-12T00:00:00.000000000', '2021-10-17T00:00:00.000000000', '2021-10-20T00:00:00.000000000', '2021-10-24T00:00:00.000000000', '2021-10-29T00:00:00.000000000', '2021-11-01T00:00:00.000000000', '2021-11-05T00:00:00.000000000', '2021-11-10T00:00:00.000000000', '2021-11-13T00:00:00.000000000', '2021-11-17T00:00:00.000000000', '2021-11-22T00:00:00.000000000', '2021-11-25T00:00:00.000000000', '2021-11-29T00:00:00.000000000', '2021-12-04T00:00:00.000000000', '2021-12-07T00:00:00.000000000', '2021-12-11T00:00:00.000000000', '2021-12-16T00:00:00.000000000', '2021-12-19T00:00:00.000000000', '2021-12-23T00:00:00.000000000', '2021-12-28T00:00:00.000000000', '2021-12-31T00:00:00.000000000', '2022-01-04T00:00:00.000000000', '2022-01-09T00:00:00.000000000', '2022-01-12T00:00:00.000000000', '2022-01-16T00:00:00.000000000', '2022-01-21T00:00:00.000000000', '2022-01-24T00:00:00.000000000', '2022-01-28T00:00:00.000000000', '2022-02-02T00:00:00.000000000', '2022-02-14T00:00:00.000000000', '2022-02-17T00:00:00.000000000', '2022-02-21T00:00:00.000000000', '2022-02-26T00:00:00.000000000', '2022-03-01T00:00:00.000000000', '2022-03-05T00:00:00.000000000', '2022-03-10T00:00:00.000000000', '2022-03-13T00:00:00.000000000', '2022-03-17T00:00:00.000000000', '2022-03-22T00:00:00.000000000', '2022-03-25T00:00:00.000000000', '2022-03-29T00:00:00.000000000', '2022-04-03T00:00:00.000000000', '2022-04-03T00:00:00.000000000', '2022-04-06T00:00:00.000000000', '2022-04-10T00:00:00.000000000', '2022-04-15T00:00:00.000000000', '2022-04-18T00:00:00.000000000', '2022-04-22T00:00:00.000000000', '2022-04-27T00:00:00.000000000', '2022-04-30T00:00:00.000000000', '2022-05-04T00:00:00.000000000', '2022-05-09T00:00:00.000000000', '2022-05-16T00:00:00.000000000', '2022-05-21T00:00:00.000000000', '2022-05-21T00:00:00.000000000', '2022-05-21T00:00:00.000000000'], dtype='datetime64[ns]') - granule_id(acq_date)<U67'S1A_IW_SLC__1SDV_20210602T12054...
- description :
- source granule ID for ASF-processed S1 RTC imagery, extracted from README files for each scene
array(['S1A_IW_SLC__1SDV_20210602T120543_20210602T120610_038161_0480FD_893F', 'S1A_IW_SLC__1SDV_20210607T121351_20210607T121418_038234_048318_9B64', 'S1A_IW_SLC__1SDV_20210610T000310_20210610T000337_038270_04841E_601A', 'S1A_IW_SLC__1SDV_20210614T120544_20210614T120611_038336_04862F_337B', 'S1A_IW_SLC__1SDV_20210619T121352_20210619T121419_038409_04884C_1DBA', 'S1A_IW_SLC__1SDV_20210622T000310_20210622T000337_038445_04895A_842E', 'S1A_IW_SLC__1SDV_20210626T120545_20210626T120612_038511_048B6C_9585', 'S1A_IW_SLC__1SDV_20210701T121352_20210701T121419_038584_048D87_F95C', 'S1A_IW_SLC__1SDV_20210701T121417_20210701T121445_038584_048D87_C79E', 'S1A_IW_SLC__1SDV_20210704T000311_20210704T000338_038620_048E99_1BAA', 'S1A_IW_SLC__1SDV_20210708T120545_20210708T120612_038686_0490AD_DD9E', 'S1A_IW_SLC__1SDV_20210713T121353_20210713T121420_038759_0492D4_EABD', 'S1A_IW_SLC__1SDV_20210713T121418_20210713T121446_038759_0492D4_47D9', 'S1A_IW_SLC__1SDV_20210713T121418_20210713T121446_038759_0492D4_47D9', 'S1A_IW_SLC__1SDV_20210716T000312_20210716T000339_038795_0493DB_53CD', 'S1A_IW_SLC__1SDV_20210720T120546_20210720T120613_038861_0495EC_C9D4', 'S1A_IW_SLC__1SDV_20210725T121354_20210725T121421_038934_04980F_1651', 'S1A_IW_SLC__1SDV_20210728T000312_20210728T000339_038970_04991E_C669', 'S1A_IW_SLC__1SDV_20210801T120547_20210801T120614_039036_049B1F_9997', 'S1A_IW_SLC__1SDV_20210806T121354_20210806T121421_039109_049D70_B0A0', ... 'S1A_IW_SLC__1SDV_20220317T120545_20220317T120612_042361_050CE5_4CCF', 'S1A_IW_SLC__1SDV_20220322T121353_20220322T121420_042434_050F59_A1B6', 'S1A_IW_SLC__1SDV_20220325T000312_20220325T000339_042470_051096_BA6F', 'S1A_IW_SLC__1SDV_20220329T120546_20220329T120613_042536_0512D7_425A', 'S1A_IW_SLC__1SDV_20220403T121418_20220403T121446_042609_05154A_51E7', 'S1A_IW_SLC__1SDV_20220403T121353_20220403T121420_042609_05154A_3FE7', 'S1A_IW_SLC__1SDV_20220406T000312_20220406T000339_042645_051682_4B40', 'S1A_IW_SLC__1SDV_20220410T120546_20220410T120613_042711_0518C0_3349', 'S1A_IW_SLC__1SDV_20220415T121353_20220415T121420_042784_051B30_EB87', 'S1A_IW_SLC__1SDV_20220418T000312_20220418T000339_042820_051C63_CAB2', 'S1A_IW_SLC__1SDV_20220422T120547_20220422T120614_042886_051E96_41EE', 'S1A_IW_SLC__1SDV_20220427T121354_20220427T121421_042959_0520EE_A42D', 'S1A_IW_SLC__1SDV_20220430T000313_20220430T000340_042995_05221E_46A1', 'S1A_IW_SLC__1SDV_20220504T120547_20220504T120614_043061_05245F_6F62', 'S1A_IW_SLC__1SDV_20220509T121355_20220509T121422_043134_0526C4_092E', 'S1A_IW_SLC__1SDV_20220516T120548_20220516T120615_043236_0529DD_42D2', 'S1A_IW_SLC__1SDV_20220521T121420_20220521T121448_043309_052C00_493D', 'S1A_IW_SLC__1SDV_20220521T121420_20220521T121448_043309_052C00_493D', 'S1A_IW_SLC__1SDV_20220521T121356_20220521T121423_043309_052C00_FE88'], dtype='<U67') - x(x)float646.194e+05 6.195e+05 ... 6.281e+05
- axis :
- X
- long_name :
- x coordinate of projection
- standard_name :
- projection_x_coordinate
- units :
- metre
array([619425., 619455., 619485., ..., 628035., 628065., 628095.])
- y(y)float643.102e+06 3.102e+06 ... 3.09e+06
- axis :
- Y
- long_name :
- y coordinate of projection
- standard_name :
- projection_y_coordinate
- units :
- metre
array([3101655., 3101625., 3101595., ..., 3089865., 3089835., 3089805.])
- spatial_ref()int640
- crs_wkt :
- PROJCS["WGS 84 / UTM zone 45N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",87],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32645"]]
- semi_major_axis :
- 6378137.0
- semi_minor_axis :
- 6356752.314245179
- inverse_flattening :
- 298.257223563
- reference_ellipsoid_name :
- WGS 84
- longitude_of_prime_meridian :
- 0.0
- prime_meridian_name :
- Greenwich
- geographic_crs_name :
- WGS 84
- horizontal_datum_name :
- World Geodetic System 1984
- projected_crs_name :
- WGS 84 / UTM zone 45N
- grid_mapping_name :
- transverse_mercator
- latitude_of_projection_origin :
- 0.0
- longitude_of_central_meridian :
- 87.0
- false_easting :
- 500000.0
- false_northing :
- 0.0
- scale_factor_at_central_meridian :
- 0.9996
- spatial_ref :
- PROJCS["WGS 84 / UTM zone 45N",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Transverse_Mercator"],PARAMETER["latitude_of_origin",0],PARAMETER["central_meridian",87],PARAMETER["scale_factor",0.9996],PARAMETER["false_easting",500000],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],AUTHORITY["EPSG","32645"]]
- GeoTransform :
- 619410.0 30.0 0.0 3101670.0 0.0 -30.0
array(0)
- sensor(acq_date)<U3'S1A' 'S1A' 'S1A' ... 'S1A' 'S1A'
array(['S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A', 'S1A'], dtype='<U3') - beam_mode(acq_date)<U2'IW' 'IW' 'IW' ... 'IW' 'IW' 'IW'
array(['IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW', 'IW'], dtype='<U2') - acquisition_time(acq_date)datetime64[ns]2022-09-27T12:14:14 ... 2022-09-...
array(['2022-09-27T12:14:14.000000000', '2022-09-27T12:13:55.000000000', '2022-09-27T00:03:12.000000000', '2022-09-27T12:14:20.000000000', '2022-09-27T00:03:13.000000000', '2022-09-27T12:05:43.000000000', '2022-09-27T00:03:15.000000000', '2022-09-27T12:05:43.000000000', '2022-09-27T12:13:56.000000000', '2022-09-27T00:03:14.000000000', '2022-09-27T12:13:53.000000000', '2022-09-27T12:05:48.000000000', '2022-09-27T12:05:46.000000000', '2022-09-27T00:03:14.000000000', '2022-09-27T00:03:12.000000000', '2022-09-27T12:05:46.000000000', '2022-09-27T12:05:49.000000000', '2022-09-27T12:14:18.000000000', '2022-09-27T00:03:11.000000000', '2022-09-27T00:03:12.000000000', '2022-09-27T12:13:53.000000000', '2022-09-27T00:03:11.000000000', '2022-09-27T12:13:54.000000000', '2022-09-27T12:13:56.000000000', '2022-09-27T12:14:18.000000000', '2022-09-27T12:05:47.000000000', '2022-09-27T00:03:15.000000000', '2022-09-27T00:03:15.000000000', '2022-09-27T12:13:52.000000000', '2022-09-27T00:03:13.000000000', '2022-09-27T12:13:50.000000000', '2022-09-27T12:13:56.000000000', '2022-09-27T12:14:17.000000000', '2022-09-27T12:13:54.000000000', '2022-09-27T12:13:53.000000000', '2022-09-27T12:05:48.000000000', '2022-09-27T00:03:10.000000000', '2022-09-27T12:05:45.000000000', '2022-09-27T12:13:53.000000000', '2022-09-27T12:05:50.000000000', ... '2022-09-27T12:13:57.000000000', '2022-09-27T00:03:12.000000000', '2022-09-27T12:05:46.000000000', '2022-09-27T12:05:49.000000000', '2022-09-27T12:05:50.000000000', '2022-09-27T12:13:53.000000000', '2022-09-27T12:13:56.000000000', '2022-09-27T00:03:14.000000000', '2022-09-27T12:05:44.000000000', '2022-09-27T12:13:57.000000000', '2022-09-27T00:03:11.000000000', '2022-09-27T12:05:46.000000000', '2022-09-27T12:05:46.000000000', '2022-09-27T12:13:49.000000000', '2022-09-27T12:05:49.000000000', '2022-09-27T12:05:45.000000000', '2022-09-27T00:03:15.000000000', '2022-09-27T12:13:57.000000000', '2022-09-27T12:14:18.000000000', '2022-09-27T00:03:12.000000000', '2022-09-27T00:03:12.000000000', '2022-09-27T00:03:16.000000000', '2022-09-27T12:13:54.000000000', '2022-09-27T12:05:50.000000000', '2022-09-27T12:13:55.000000000', '2022-09-27T12:05:49.000000000', '2022-09-27T12:05:48.000000000', '2022-09-27T12:05:48.000000000', '2022-09-27T12:05:47.000000000', '2022-09-27T12:13:55.000000000', '2022-09-27T12:13:51.000000000', '2022-09-27T12:13:54.000000000', '2022-09-27T00:03:09.000000000', '2022-09-27T00:03:11.000000000', '2022-09-27T00:03:13.000000000', '2022-09-27T12:14:14.000000000', '2022-09-27T12:13:57.000000000', '2022-09-27T00:03:12.000000000'], dtype='datetime64[ns]') - polarisation_type(acq_date)<U2'DV' 'DV' 'DV' ... 'DV' 'DV' 'DV'
array(['DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV', 'DV'], dtype='<U2') - orbit_type(acq_date)<U1'P' 'P' 'P' 'P' ... 'P' 'P' 'P' 'P'
array(['P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P', 'P'], dtype='<U1') - terrain_correction_pixel_spacing(acq_date)<U5'RTC30' 'RTC30' ... 'RTC30' 'RTC30'
array(['RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30', 'RTC30'], dtype='<U5') - output_format(acq_date)<U1'g' 'g' 'g' 'g' ... 'g' 'g' 'g' 'g'
array(['g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g', 'g'], dtype='<U1') - output_type(acq_date)<U1'p' 'p' 'p' 'p' ... 'p' 'p' 'p' 'p'
array(['p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p', 'p'], dtype='<U1') - masked(acq_date)<U1'u' 'u' 'u' 'u' ... 'u' 'u' 'u' 'u'
array(['u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u', 'u'], dtype='<U1') - filtered(acq_date)<U1'n' 'n' 'n' 'n' ... 'n' 'n' 'n' 'n'
array(['n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n', 'n'], dtype='<U1') - area(acq_date)<U1'e' 'e' 'e' 'e' ... 'e' 'e' 'e' 'e'
array(['e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e', 'e'], dtype='<U1') - product_id(acq_date)<U4'1424' 'ABA0' ... '01F4' 'BD36'
array(['1424', 'ABA0', 'E61E', 'E5B6', '5569', '71C9', 'B942', 'DCE1', 'CA1B', '6581', '1DFD', '5413', '22BD', 'BDC1', '01B2', '9849', 'CFFC', '57F2', 'D4E6', 'B7B0', '3770', '52BB', '0579', 'AA68', '24B8', '5FF4', '5F4E', '199D', '0031', 'D082', 'E74B', 'F7ED', '65E0', '528F', '51E7', 'F5AA', 'AC38', '772E', 'F0A6', '066E', '5844', '8CD8', '6340', 'D35A', '056C', '6E6F', '0CAD', '6270', '8A4F', 'BEAB', '54B1', '8F7B', 'F9A9', 'AA59', '57E1', '81EB', '250E', '3C39', '3A2A', '9331', 'C14C', 'F1DE', '106F', 'FD00', '5D96', '971C', 'F573', 'C890', '68EA', '83D7', 'E113', 'D846', 'F4B3', 'BB26', '485C', 'B409', '6683', '6153', '832B', '7418', '378D', '33F5', '1139', 'B661', '9A2F', '6943', '5BAD', 'FA4F', '01F4', 'BD36'], dtype='<U4') - acq_hour(acq_date)int6412 12 0 12 0 12 0 ... 0 0 0 12 12 0
array([12, 12, 0, 12, 0, 12, 0, 12, 12, 0, 12, 12, 12, 0, 0, 12, 12, 12, 0, 0, 12, 0, 12, 12, 12, 12, 0, 0, 12, 0, 12, 12, 12, 12, 12, 12, 0, 12, 12, 12, 12, 0, 12, 0, 12, 12, 12, 12, 12, 12, 0, 12, 12, 0, 12, 12, 12, 12, 12, 0, 12, 12, 0, 12, 12, 12, 12, 12, 0, 12, 12, 0, 0, 0, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 0, 0, 0, 12, 12, 0]) - orbital_dir(acq_date)<U4'desc' 'desc' ... 'desc' 'asc'
array(['desc', 'desc', 'asc', 'desc', 'asc', 'desc', 'asc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'desc', 'desc', 'asc', 'asc', 'asc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'desc', 'asc', 'asc', 'asc', 'desc', 'desc', 'asc'], dtype='<U4') - data_take_id(acq_date)<U6'0480FD' '048318' ... '052C00'
- ID of data take from SAR acquisition :
- ID data take
array(['0480FD', '048318', '04841E', '04862F', '04884C', '04895A', '048B6C', '048D87', '048D87', '048E99', '0490AD', '0492D4', '0492D4', '0492D4', '0493DB', '0495EC', '04980F', '04991E', '049B1F', '049D70', '049EAC', '04A0FF', '04A383', '04A4B9', '04A6FC', '04AAB3', '04AD0D', '04AF85', '04B0C4', '04B2FB', '04B566', '04B6AB', '04B8FF', '04BCB8', '04BF12', '04C195', '04C2D6', '04C52E', '04C7A8', '04C8E9', '04CB46', '04CDC8', '04CEFF', '04D14F', '04D3CC', '04D50E', '04D761', '04D9E8', '04DB21', '04DD71', '04DFDD', '04E110', '04E340', '04E5AB', '04E6D7', '04E931', '04EB9B', '04ECCD', '04EEE8', '04F156', '04F294', '04F4D6', '04F754', '04FD6F', '04FEB2', '050108', '050376', '0504AF', '0506F7', '050962', '050AA2', '050CE5', '050F59', '051096', '0512D7', '05154A', '05154A', '051682', '0518C0', '051B30', '051C63', '051E96', '0520EE', '05221E', '05245F', '0526C4', '0529DD', '052C00', '052C00', '052C00'], dtype='<U6')
- vv(acq_date, y, x)float32dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
- scale_factor :
- 1.0
- add_offset :
- 0.0
- _FillValue :
- 0.0
Array Chunk Bytes 39.43 MiB 448.59 kiB Shape (90, 396, 290) (1, 396, 290) Count 17 Graph Layers 90 Chunks Type float32 numpy.ndarray - vh(acq_date, y, x)float32dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
- scale_factor :
- 1.0
- add_offset :
- 0.0
- _FillValue :
- 0.0
Array Chunk Bytes 39.43 MiB 448.59 kiB Shape (90, 396, 290) (1, 396, 290) Count 27 Graph Layers 90 Chunks Type float32 numpy.ndarray - ls(acq_date, y, x)float64dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
- scale_factor :
- 1.0
- add_offset :
- 0.0
- _FillValue :
- 0
Array Chunk Bytes 78.85 MiB 897.19 kiB Shape (90, 396, 290) (1, 396, 290) Count 31 Graph Layers 90 Chunks Type float64 numpy.ndarray
asf_desc = asf_subset.where(asf_subset['orbital_dir'] == 'desc', drop=True)
asf_asc = asf_subset.where(asf_subset['orbital_dir'] == 'asc', drop=True)
#asf_asc.vv.mean(dim=['x','y']).plot(marker='o', linestyle='None')
pc_desc = pc_subset.where(pc_subset['sat:orbit_state'] == 'descending', drop=True)
pc_asc = pc_subset.where(pc_subset['sat:orbit_state'] == 'ascending', drop=True)
fig, axs = plt.subplots(ncols =2 ,figsize=(16,8))
power_to_db(asf_desc.vv.mean(dim=['x','y'])).plot(ax=axs[0], color = 'blue', linestyle='None', marker='+', markersize=5, label = 'ASF VV desc');
power_to_db(pc_desc.sel(band='vv').mean(dim=['x','y'])).plot(ax=axs[0], color = 'red', linestyle='None', marker = '+', markersize=5, label = 'PC VV desc');
power_to_db(asf_asc.vv.mean(dim=['x','y'])).plot(ax=axs[0], color = 'blue', linestyle='None', marker='o', markersize=5, label = 'ASF VV asc');
power_to_db(pc_asc.sel(band='vv').mean(dim=['x','y'])).plot(ax=axs[0], color = 'red', linestyle='None', marker = 'o', markersize=5, label = 'PC VV asc');
axs[0].legend(loc = 'lower right')
axs[0].set_xlabel('Acquisition date')
axs[0].set_ylabel('Backscatter (sigma-nought) (dB)')
axs[0].set_title('VV')
power_to_db(asf_desc.vh.mean(dim=['x','y'])).plot(ax=axs[1], color = 'blue', linestyle='None', marker='+', markersize=5, label = 'ASF VH desc');
power_to_db(pc_desc.sel(band='vh').mean(dim=['x','y'])).plot(ax=axs[1], color = 'red', linestyle='None', marker = '+', markersize=5, label = 'PC VH desc');
power_to_db(asf_asc.vh.mean(dim=['x','y'])).plot(ax=axs[1], color = 'blue', linestyle='None', marker='o', markersize=5, label = 'ASF VH asc');
power_to_db(pc_asc.sel(band='vh').mean(dim=['x','y'])).plot(ax=axs[1], color = 'red', linestyle='None', marker = 'o', markersize=5, label = 'PC VH asc');
axs[1].legend(loc = 'lower right')
axs[1].set_xlabel('Acquisition date')
axs[1].set_ylabel('Backscatter (sigma-nought) (dB)')
axs[1].set_title('VH')
fig.suptitle('Comparison of backscatter values from ASF-processed and PC-processed Sentinel-1 RTC imagery');
Text(0.5, 0.98, 'Comparison of backscatter values from ASF-processed and PC-processed Sentinel-1 RTC imagery')