ASF GDAL VRT approach#
This notebook demonstrates working with Sentinel-1 RTC imagery that has been processed on the ASF On-Demand server and downloaded locally.
The access point for data in this notebook is a directory containing un-zipped directories of RTC scenes.
On reading the data#
Two approaches were tested for reading in the large stack of Sentinel-1 time series data: using GDAL VRT objects and xarray’s open_mfdataset(). The VRT approach ended up being the most appropriate for this situation but you can also view the xr.open_mfdataset() in the appendix if you’re interested. Here’s a brief description of the pros and cons of each approach.
xr.open_mfdataset()#
This function allows the user to read in a large number of files into a single xarray object in one step. This can save a huge amount of time reading in files and reorganizing xarray objects.
Pros#
Built-in xarray functionality
Files can be distributed - read from directory or list of filepaths
Large degree of customization via preprocess argument
Dask integration
Cons#
Static chunking strategy through stack
Not well-suited for files with non-uniform spatial footprints
Cannot lazily perform clipping operations
GDAL Virtual Raster Format (VRT)#
VRT is short for GDAL Virtual Format. VRT objects create an xml file from a list of geotiff files. The xml contains a mapping of all of the specified raster files so that we essentially have the spatial information that would have been used to create the full data object, and all of the information we need for a clip.
Pros#
Can handle a mismatch of grids within the file stack
Xml structure can store a large amount of information in a lightweight way
Flexible, can add files into the same or separate bands of VRT object
Cons#
Less customization than
xr.open_mfdataset()Lose some structure (files are renamed Band 1,2,3…)
Lose metadata
Because of the size and some characteristics of this dataset, it was computationally expensive to read the geotiff data in directly using xarray methods such as xarray.open_mfdataset. Instead, the GDAL VRT approach was able to handle the grid structures within the stack of files and did not encounter the memory issues we had when trying to use xr.open_mfdataset() with dask.
The GDAL VRT approach let’s you read in the data stack with less computational expense, but because of how the data is represented, some metadata is lost and must be re-structured. We will go through those steps here, with explanations of the xarray tools we use along the way.
Learning goals#
Techniques (xarray, python and general):
Build gdal VRT objects
Read in data using
rioxarray.open_rasterio()Clip raster object by a vector using
rioxarray.clip()Indexing and selecting xarray objects by dimensional and non-dimensional coordinates
Formatting time-aware xarray objects
High-level science goals:
Organize large set of geotiff files stored locally
Use VRT objects to create xarray objects from large data stacks
Extract and organize metadata stored in filenames and text files
Handle
nodatavalues
Other useful resources#
These are resources that contain additional examples and discussion of the content in this notebook and more.
How do I… this is very helpful!
Xarray High-level computational patterns discussion of concepts and associated code examples
Parallel computing with dask Xarray tutorial demonstrating wrapping of dask arrays
ASF Data Access#
You can download the RTC-processed backscatter time series here
Software and setup#
%xmode minimal
import os
import xarray as xr
import rioxarray as rio
import geopandas as gpd
from datetime import datetime
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import dask
import markdown
import pathlib
from pathlib import Path
import time
import glob
%load_ext watermark
%watermark -iv
pandas : 2.2.0
xarray : 2024.1.1
markdown : 3.5.2
numpy : 1.26.3
geopandas : 0.14.3
matplotlib: 3.8.2
rioxarray : 0.15.1
dask : 2024.1.1
Utility Functions#
#we'll use this function to get bounding box coordinates from a list of points
def points2coords(pt_ls): #should be [xmin, ymin, xmax, ymax]
coords_ls = [(pt_ls[0], pt_ls[1]), (pt_ls[0], pt_ls[3]),
(pt_ls[2], pt_ls[3]), (pt_ls[2], pt_ls[1]),
(pt_ls[0], pt_ls[1])]
return coords_ls
def power_to_db(input_arr):
return (10*np.log10(np.abs(input_arr)))
Read in vector data#
We will use this vector data object later but we will read it in as a geopandas.GeoDataFrame object now. It is called PC aoi because the GeoJSON file was created off of the spatial extent of the SAR data we will access from Microsoft Planetary Computer (PC), in order to have comparable datasets.
pc_aoi = gpd.read_file('https://github.com/e-marshall/sentinel1_rtc/raw/main/hma_rtc_aoi.geojson')
pc_aoi
| geometry | |
|---|---|
| 0 | POLYGON ((619420.000 3089790.000, 628100.000 3... |
pc_aoi.plot(edgecolor='blue', facecolor='None')
<Axes: >
Organizing Sentinel-1 data to create VRT objects.#
Setup string variables to access filenames + filepaths. Currently, the file structure is organized so that each scene has its own sub-directory within asf_rtcs. Within each sub-directory are several files - we want to extract the tif files containing RTC imagery for the VV and VH polarizations for each scene.
The function extract_tif_fnames() takes a path to the directory containing the sub-directories for all scenes and returns a lists of filenames for VV polarization GeoTIFF files, VH polarization GeoTIFF files and the layover-shadow mask GeoTIFF files. Separately, it returns a list of filenames for the associated README files which will be used to extract metadata later on.
Note
If you’re following along in your own python environment, replace the string below named dir_path_all with the path to the directory containing the RTC data on your local machine, then execute extract_tif_fnames().
dir_path_all = '/home/emmamarshall/Desktop/siparcs/asf_rtc_data/asf_rtcs/'
scenes_ls = os.listdir(dir_path_all)
def extract_tif_fnames(scene_path):
''' return a list of files associated with a single S1 scene'''
scene_files_ls = os.listdir(dir_path_all + scene_path)
#make object for readme file
rm = [file for file in scene_files_ls if file.endswith('README.md.txt')]
scene_files_vh = [fname for fname in scene_files_ls if fname.endswith('_VH.tif')]
scene_files_vv = [fname for fname in scene_files_ls if fname.endswith('_VV.tif')]
scene_files_ls = [fname for fname in scene_files_ls if fname.endswith('_ls_map.tif')]
return scene_files_vv, scene_files_vh, scene_files_ls, rm
Every scene will have the following four elements:
VH band geotiff
VV band geotiff
Layover-shadow map geotiff
README text file
extract_tif_fnames(scenes_ls[0])
(['S1A_IW_20210821T000314_DVP_RTC30_G_gpuned_748F_VV.tif'],
['S1A_IW_20210821T000314_DVP_RTC30_G_gpuned_748F_VH.tif'],
['S1A_IW_20210821T000314_DVP_RTC30_G_gpuned_748F_ls_map.tif'],
['S1A_IW_20210821T000314_DVP_RTC30_G_gpuned_748F.README.md.txt'])
Now we need to attach the filenames to the full path to each file. We will end up with a list of full paths to VV and VH band imagery, layover shadow map and README files.
fpaths_vv, fpaths_vh, fpaths_ls, fpaths_rm = [],[],[], []
for element in range(len(scenes_ls)):
good_files = extract_tif_fnames(scenes_ls[element])
path_vh = dir_path_all + scenes_ls[element] + '/' + good_files[0][0]
path_vv = dir_path_all + scenes_ls[element] + '/' + good_files[1][0]
path_ls = dir_path_all + scenes_ls[element] + '/' + good_files[2][0]
path_readme = dir_path_all + scenes_ls[element] + '/' + good_files[3][0]
fpaths_vv.append(path_vv)
fpaths_vh.append(path_vh)
fpaths_ls.append(path_ls)
fpaths_rm.append(path_readme)
Create VRT objects#
We will be using the gdalbuildvrt command. You can find out more about it here. This command can make a VRT that either tiles the listed files into a large mosaic, or places them each in a separate band of the VRT. Because we are dealing with a temporal stack of images we want to use the -separate flag to place each file into a band of the VRT.
Because we are making a VRT from a large number of files, we’ll supply a text file with the full path for each file rather than include these at the end of the command line. We’ll use the same lists of filepaths that we constructed above and will write them to text files that we pass to the gdalbuildvrt command.
Note
If you are following along on your machine, be sure to replace the file paths in the below cell with the location where you would like the text files to be written.
with open('/home/emmamarshall/Desktop/data/written_data/s1_vv_fpaths.txt','w') as fp:
for item in fpaths_vv:
#write each item on a new line
fp.write('%s\n' % item)
with open('/home/emmamarshall/Desktop/data/written_data/s1_vh_fpaths.txt','w') as fp:
for item in fpaths_vh:
fp.write('%s\n' % item)
with open('/home/emmamarshall/Desktop/data/written_data/s1_ls_fpaths.txt','w') as fp:
for item in fpaths_ls:
fp.write('%s\n' % item)
Next, we will use gdal command line tools to create the VRT objects:
Note
If you are following along on your machine, be sure to replace the file paths in the below cell with the location of the .txt files written in the step above.
# syntax for gdalbuildvrt
#`gdalbuildvrt -separate -input_file_list /path/to/my_list.txt vrt_fname.vrt` </br>
# commands used to creat vrt files
!gdalbuildvrt -separate -input_file_list /home/emmamarshall/Desktop/data/written_data/s1_vv_fpaths.txt s1_stackVV.vrt
!gdalbuildvrt -separate -input_file_list /home/emmamarshall/Desktop/data/written_data/s1_vh_fpaths.txt s1_stackVH.vrt
!gdalbuildvrt -separate -input_file_list /home/emmamarshall/Desktop/data/written_data/s1_ls_fpaths.txt s1_stackLS.vrt
0...10...20...30...40...50...60...70...80...90...100 - done.
0...10...20...30...40...50...60...70...80...90...100 - done.
0...10...20...30...40...50...60...70...80...90...100 - done.
Read in the VRT files using rioxarray.open_rasterio():
vrt_vv = rio.open_rasterio('/home/emmamarshall/Desktop/siparcs/sentinel1_rtc/s1_stackVV.vrt', chunks='auto').squeeze()
vrt_vh = rio.open_rasterio('/home/emmamarshall/Desktop/siparcs/sentinel1_rtc/s1_stackVH.vrt', chunks='auto').squeeze()
vrt_ls = rio.open_rasterio('/home/emmamarshall/Desktop/siparcs/sentinel1_rtc/s1_stackLS.vrt', chunks='auto').squeeze()
vrt_vv
<xarray.DataArray (band: 103, y: 13379, x: 17452)>
dask.array<open_rasterio-b10d33d8a4b01aa9049ac0a5fdaa0889<this-array>, shape=(103, 13379, 17452), dtype=float32, chunksize=(1, 5760, 5760), chunktype=numpy.ndarray>
Coordinates:
* band (band) int64 1 2 3 4 5 6 7 8 9 ... 96 97 98 99 100 101 102 103
* x (x) float64 3.833e+05 3.833e+05 ... 9.068e+05 9.068e+05
* y (y) float64 3.309e+06 3.309e+06 ... 2.907e+06 2.907e+06
spatial_ref int64 0
Attributes:
_FillValue: 0.0
scale_factor: 1.0
add_offset: 0.0Building the VRT object assigns every object in the .txt file to a different band. In doing this, we lose the metadata that is associated with the files. We need to extract metadata from the file name and attach it to the xarray objects that we read in from VRTs.
Extract metadata#
The following function is very similar to the preprocess() function that is passed when calling xr.open_mfdataset(). There is an example of this in the appendix notebook. It extracts metadata contained in the filename and organizes it as a dictionary that can be attached to an xarray object.
def extract_metadata_attrs(inputfname):
#vv_fn = da_orig.encoding['source'][113:]
vv_fn = inputfname[108:]
sensor = vv_fn[0:3]
beam_mode = vv_fn[4:6]
acq_date_full = vv_fn[7:22]
acq_datetime = pd.to_datetime(acq_date_full, format='%Y%m%dT%H%M%S')
pol_type = vv_fn[23:25]
orbit_type = vv_fn[25:26]
orbit_type = vv_fn[26:27] #Precise (P), Restituted (R), or Original Predicted (O)
terrain_correction_pixel_spacing = vv_fn[27:32] #Terrain Correction Pixel Spacing
rtc_alg = vv_fn[33:34] #Software Package Used: GAMMA (G)
output = vv_fn[35] # Gamma-0 (g) or Sigma-0 (s) Output
output_type = vv_fn[36] #Power (p) or Decibel (d) or Amplitude (a) Output
masked = vv_fn[37] #Unmasked (u) or Water Masked (w)
filtered = vv_fn[38] # Not Filtered (n) or Filtered (f)
area = vv_fn[39] # Entire Area (e) or Clipped Area (c)
tbd = vv_fn[40] #Dead Reckoning (d) or DEM Matching (m)
# product_id = vv_fn[56:62] #Product ID
product_id = vv_fn[42:46]
attrs_dict = { 'sensor': sensor,
'beam_mode':beam_mode,
'acquisition_datetime': acq_datetime,
'polarisation_type': pol_type,
# 'primary_polarisation': primary_pol,
'orbit_type': orbit_type,
'terrain_correction_pixel_spacing' : terrain_correction_pixel_spacing,
'output_format': output,
'output_type': output_type,
'masked' : masked,
'filtered':filtered,
'area':area,
'product_id': product_id
}
return attrs_dict
The acquisition dates extracted from the VV, VH and L-S geotiff files should be identical, but we read in all three here just to confirm:
acq_dates_vh = [extract_metadata_attrs(fpaths_vh[file])['acquisition_datetime'].strftime('%m/%d/%YT%H%M%S') for file in range(len(fpaths_vh))]
acq_dates_vv = [extract_metadata_attrs(fpaths_vv[file])['acquisition_datetime'].strftime('%m/%d/%YT%H%M%S') for file in range(len(fpaths_vv))]
acq_dates_ls = [extract_metadata_attrs(fpaths_ls[file])['acquisition_datetime'].strftime('%m/%d/%YT%H%M%S') for file in range(len(fpaths_vv))]
acq_dates_vh[:5]
['08/21/2021T000314',
'01/16/2022T120547',
'07/08/2021T120545',
'04/22/2022T120547',
'04/27/2022T121354']
The acquisition dates for VV, VH and LS should be the same. As a check, you can confirm this by creating both objects and comparing them.
acq_dates_vh == acq_dates_vv == acq_dates_ls
True
We can now assign the list of acquisition dates as coordinates to the xarray objects. Use pd.to_datetime() to do this as time-aware coordinate values.
vrt_vv = vrt_vv.assign_coords({'band':pd.to_datetime(acq_dates_vv, format='%m/%d/%YT%H%M%S')})
vrt_vh = vrt_vh.assign_coords({'band':pd.to_datetime(acq_dates_vh, format='%m/%d/%YT%H%M%S')})
vrt_ls = vrt_ls.assign_coords({'band':pd.to_datetime(acq_dates_ls, format='%m/%d/%YT%H%M%S')})
Merge the three xr.DataArrays together into an xr.Dataset.
vrt_merge = xr.Dataset({'vv':vrt_vv,
'vh':vrt_vh,
'ls':vrt_ls}).rename({'band':'acq_date'})
vrt_merge
<xarray.Dataset>
Dimensions: (x: 17452, y: 13379, acq_date: 103)
Coordinates:
* x (x) float64 3.833e+05 3.833e+05 ... 9.068e+05 9.068e+05
* y (y) float64 3.309e+06 3.309e+06 ... 2.907e+06 2.907e+06
spatial_ref int64 0
* acq_date (acq_date) datetime64[ns] 2021-08-21T00:03:14 ... 2022-04-06...
Data variables:
vv (acq_date, y, x) float32 dask.array<chunksize=(1, 5760, 5760), meta=np.ndarray>
vh (acq_date, y, x) float32 dask.array<chunksize=(1, 5760, 5760), meta=np.ndarray>
ls (acq_date, y, x) uint8 dask.array<chunksize=(1, 11520, 11520), meta=np.ndarray>The files are not organized by time in the directory, so the files are not read in in temporal order when we create the VRT and xr.Dataset objects. Once we have attached the temporal metadata to the xarray objects composed from VRT, we can use Dataset.sortby() to organize the dataset along time time dimension:
vrt_merge = vrt_merge.sortby(vrt_merge.acq_date)
vrt_merge
<xarray.Dataset>
Dimensions: (x: 17452, y: 13379, acq_date: 103)
Coordinates:
* x (x) float64 3.833e+05 3.833e+05 ... 9.068e+05 9.068e+05
* y (y) float64 3.309e+06 3.309e+06 ... 2.907e+06 2.907e+06
spatial_ref int64 0
* acq_date (acq_date) datetime64[ns] 2021-05-02T12:14:14 ... 2022-05-21...
Data variables:
vv (acq_date, y, x) float32 dask.array<chunksize=(1, 5760, 5760), meta=np.ndarray>
vh (acq_date, y, x) float32 dask.array<chunksize=(1, 5760, 5760), meta=np.ndarray>
ls (acq_date, y, x) uint8 dask.array<chunksize=(1, 11520, 11520), meta=np.ndarray>Taking a look at chunking#
If you take a look at the chunking you will see that the entire object has a shape (103, 13379, 17452) and that each chunk is (1, 5760, 5760). This breaks the full array (~ 89 GB) into 1,236 chunks that are about 127 MB each. We can also see that chunking keeps each time step intact which is optimal for time series data. If you are interested in an example of inefficient chunking, you can check out the example notebook in the appendix. In this case, because of the internal structure of the data and the characteristics of the time series stack, various chunking strategies produced either too few (103) or too many (317,240) chunks with complicated structures that led to memory blow-ups when trying to compute. The difficulty we encountered trying to structure the data using xr.open_mfdataset() led us to use the VRT approach in this notebook but xr.open_mfdataset() is still a very useful tool if your data is a good fit.
Chunking is an important aspect of how dask works. You want the chunking strategy to match the structure of the data (ie. internal tiling of the data, if your data is stored locally you want chunks to match the storage structure) without having too many chunks (this will cause unnecessary communication among workers) or too few chunks (this will lead to large chunk sizes and slower processing). There are helpful explanations here and here.
When chunking is set to auto (the case here), the optimal chunk size will be selected for each dimension (if specified individually) or all dimensions. Read more about chunking here.
Clip by vector data#
Clip the full object by the same AOI as above. We will use the rioxarray.clip() method.
vrt_clip = vrt_merge.rio.clip(pc_aoi.geometry, pc_aoi.crs)
vrt_clip
<xarray.Dataset>
Dimensions: (x: 290, y: 396, acq_date: 103)
Coordinates:
* x (x) float64 6.194e+05 6.195e+05 ... 6.281e+05 6.281e+05
* y (y) float64 3.102e+06 3.102e+06 3.102e+06 ... 3.09e+06 3.09e+06
* acq_date (acq_date) datetime64[ns] 2021-05-02T12:14:14 ... 2022-05-21...
spatial_ref int64 0
Data variables:
vv (acq_date, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
vh (acq_date, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
ls (acq_date, y, x) uint8 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>Merge metadata#
We had important metadata stored in the individual filenames of each Sentinel-1 scene that was dropped when we converted them to VRT objects. Let’s use the original filenames to extract the metadata and assign the data as coordinates of our formatted xarray object.
First here is what the metadata looks like for a single file
extract_metadata_attrs(fpaths_vv[0])
{'sensor': 'S1A',
'beam_mode': 'IW',
'acquisition_datetime': Timestamp('2021-08-21 00:03:14'),
'polarisation_type': 'DV',
'orbit_type': '_',
'terrain_correction_pixel_spacing': 'RTC30',
'output_format': 'g',
'output_type': 'p',
'masked': 'u',
'filtered': 'n',
'area': 'e',
'product_id': '748F'}
meta_attrs_ls_vv = [extract_metadata_attrs(fpaths_vv[file]) for file in range(len(fpaths_vv))]
def stacked_meta_tuple(input_ls):
'''takes a list of dictionaries where each dict is metadata for a given time step,
returns a tuple. input list is re-organized to a tuple where each [0] values is a metadata category (ie. sensor) and
[1] is a time series of the categories values over time (ie. S1A, S1A, S1A ...)'''
meta_dict = input_ls[0]
ticker = 0
attrs_dicts, keys_ls = [],[]
for key in meta_dict:
if key == 'acquisition_datetime':
pass
else:
key_dict = {f'{key}':[input_ls[ticker][key] for ticker in range(len(input_ls))]}
ticker+=1
attrs_dicts.append(key_dict)
keys_ls.append(key)
full_tuple = tuple(zip(keys_ls, attrs_dicts))
return full_tuple
def apply_meta_coords(input_xr, input_tuple):
'''takes an input xarray object and a tuple of metadata created from the above fn.
returns xr object with metadata from tuple applied
'''
out_xr = xr.Dataset(
input_xr.data_vars,
coords = {'x': input_xr.x.data,
'y': input_xr.y.data,
# 'acq_date': input_xr.acq_date.data,
}
)
# now apply metadata coords
for element in range(len(input_tuple)):
key = input_tuple[element][0]
coord = list(input_tuple[element][1].values())[0]
out_xr.coords[f'{key}'] = ('acq_date', coord)
# print(key)
# print(out_xr.coords.values)
return out_xr
meta_tuple = stacked_meta_tuple(meta_attrs_ls_vv)
vrt_full = apply_meta_coords(vrt_clip, meta_tuple)
vrt_full
<xarray.Dataset>
Dimensions: (x: 290, y: 396, acq_date: 103)
Coordinates: (12/15)
* x (x) float64 6.194e+05 ... 6.281e+05
* y (y) float64 3.102e+06 ... 3.09e+06
* acq_date (acq_date) datetime64[ns] 2021-05-02T12...
spatial_ref int64 0
sensor (acq_date) <U3 'S1A' 'S1A' ... 'S1A' 'S1A'
beam_mode (acq_date) <U2 'IW' 'IW' ... 'IW' 'IW'
... ...
output_format (acq_date) <U1 'g' 'g' 'g' ... 'g' 'g' 'g'
output_type (acq_date) <U1 'p' 'p' 'p' ... 'p' 'p' 'p'
masked (acq_date) <U1 'u' 'u' 'u' ... 'u' 'u' 'u'
filtered (acq_date) <U1 'n' 'n' 'n' ... 'n' 'n' 'n'
area (acq_date) <U1 'e' 'e' 'e' ... 'e' 'e' 'e'
product_id (acq_date) <U4 '748F' '0D1E' ... 'BD36'
Data variables:
vv (acq_date, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
vh (acq_date, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
ls (acq_date, y, x) uint8 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>Add granule ID as another non-dimensional coordinate#
Extract SLC granule ID from readme#
The original granule ID associated with the Sentinel-1 data product published by the European Space Agency contains metadata that it would be helpful for us to attach to our xarray objects. The README file that is associated with every scene contains the source granule used to generate that product.
The following two functions will be used to extract the source granule ID from each README file then to attach the ID as a coordinate to the xarray object containing the SAR imagery.
Using the granule_da object created earlier
def extract_granule_id(filepath):
''' takes a filepath to the readme associated with an S1 scene and returns the source granule id used to generate the RTC imagery'''
md = markdown.Markdown(extensions=['meta'])
data = pathlib.Path(filepath).read_text()
#this text precedes granule ID in readme
gran_str = 'The source granule used to generate the products contained in this folder is:\n'
split = data.split(gran_str)
#isolate the granule id
gran_id = split[1][:67]
return gran_id
Construct granule ID coordinate#
def make_granule_coord(readme_fpaths_ls):
'''takes a list of the filepaths to every read me, extracts the granule ID,
extracts acq date for each granule ID, organizes this as an array that
can be assigned as a coord to an xarray object'''
granule_ls = [extract_granule_id(readme_fpaths_ls[element]) for element in range(len(readme_fpaths_ls))]
acq_date = [pd.to_datetime(granule[17:25]) for granule in granule_ls]
granule_da = xr.DataArray(data = granule_ls,
dims = ['acq_date'],
coords = {'acq_date':acq_date},
attrs = {'description': 'source granule ID for ASF-processed S1 RTC imagery, extracted from README files for each scene'},
name = 'granule_id')
granule_da = granule_da.sortby(granule_da.acq_date)
return granule_da
The object below (granule_da) is a 2-dimensional xr.DataArray that contains an acquisition date coordinate dimension and an array of Sentinel-1 granule IDs. It will be used later when we are merging metadata with our imagery object.
granule_da = make_granule_coord(fpaths_rm)
granule_da
<xarray.DataArray 'granule_id' (acq_date: 103)>
array(['S1A_IW_SLC__1SDV_20210502T121414_20210502T121441_037709_047321_900F',
'S1A_IW_SLC__1SDV_20210505T000307_20210505T000334_037745_047463_4DEF',
'S1A_IW_SLC__1SDV_20210509T120542_20210509T120609_037811_047676_9EBD',
'S1A_IW_SLC__1SDV_20210514T121349_20210514T121416_037884_047898_4702',
'S1A_IW_SLC__1SDV_20210514T121414_20210514T121442_037884_047898_FE6F',
'S1A_IW_SLC__1SDV_20210517T000308_20210517T000335_037920_0479A9_2A18',
'S1A_IW_SLC__1SDV_20210521T120543_20210521T120609_037986_047BBD_1CA0',
'S1A_IW_SLC__1SDV_20210526T121350_20210526T121417_038059_047DE5_0E67',
'S1A_IW_SLC__1SDV_20210529T000309_20210529T000336_038095_047EF4_5449',
'S1A_IW_SLC__1SDV_20210602T120543_20210602T120610_038161_0480FD_893F',
'S1A_IW_SLC__1SDV_20210607T121351_20210607T121418_038234_048318_9B64',
'S1A_IW_SLC__1SDV_20210610T000310_20210610T000337_038270_04841E_601A',
'S1A_IW_SLC__1SDV_20210614T120544_20210614T120611_038336_04862F_337B',
'S1A_IW_SLC__1SDV_20210619T121352_20210619T121419_038409_04884C_1DBA',
'S1A_IW_SLC__1SDV_20210622T000310_20210622T000337_038445_04895A_842E',
'S1A_IW_SLC__1SDV_20210626T120545_20210626T120612_038511_048B6C_9585',
'S1A_IW_SLC__1SDV_20210701T121352_20210701T121419_038584_048D87_F95C',
'S1A_IW_SLC__1SDV_20210701T121417_20210701T121445_038584_048D87_C79E',
'S1A_IW_SLC__1SDV_20210704T000311_20210704T000338_038620_048E99_1BAA',
'S1A_IW_SLC__1SDV_20210708T120545_20210708T120612_038686_0490AD_DD9E',
...
'S1A_IW_SLC__1SDV_20220317T120545_20220317T120612_042361_050CE5_4CCF',
'S1A_IW_SLC__1SDV_20220322T121353_20220322T121420_042434_050F59_A1B6',
'S1A_IW_SLC__1SDV_20220325T000312_20220325T000339_042470_051096_BA6F',
'S1A_IW_SLC__1SDV_20220329T120546_20220329T120613_042536_0512D7_425A',
'S1A_IW_SLC__1SDV_20220403T121418_20220403T121446_042609_05154A_51E7',
'S1A_IW_SLC__1SDV_20220403T121353_20220403T121420_042609_05154A_3FE7',
'S1A_IW_SLC__1SDV_20220406T000312_20220406T000339_042645_051682_4B40',
'S1A_IW_SLC__1SDV_20220410T120546_20220410T120613_042711_0518C0_3349',
'S1A_IW_SLC__1SDV_20220415T121353_20220415T121420_042784_051B30_EB87',
'S1A_IW_SLC__1SDV_20220418T000312_20220418T000339_042820_051C63_CAB2',
'S1A_IW_SLC__1SDV_20220422T120547_20220422T120614_042886_051E96_41EE',
'S1A_IW_SLC__1SDV_20220427T121354_20220427T121421_042959_0520EE_A42D',
'S1A_IW_SLC__1SDV_20220430T000313_20220430T000340_042995_05221E_46A1',
'S1A_IW_SLC__1SDV_20220504T120547_20220504T120614_043061_05245F_6F62',
'S1A_IW_SLC__1SDV_20220509T121355_20220509T121422_043134_0526C4_092E',
'S1A_IW_SLC__1SDV_20220516T120548_20220516T120615_043236_0529DD_42D2',
'S1A_IW_SLC__1SDV_20220521T121420_20220521T121448_043309_052C00_493D',
'S1A_IW_SLC__1SDV_20220521T121420_20220521T121448_043309_052C00_493D',
'S1A_IW_SLC__1SDV_20220521T121356_20220521T121423_043309_052C00_FE88'],
dtype='<U67')
Coordinates:
* acq_date (acq_date) datetime64[ns] 2021-05-02 2021-05-05 ... 2022-05-21
Attributes:
description: source granule ID for ASF-processed S1 RTC imagery, extract...Each line above is the full granule ID for the Sentinel-1 acquisition from 6/2/2021. You can read about the Sentinel-1 file naming convention here, but for now we are interested in the 6-digit mission data take ID.
Example from the first line of granule_da:
S1A_IW_SLC__1SDV_20210502T121414_20210502T121441_037709_047321_900F
The acquisition date for this image was 05/02/2021 and the mission data take ID is 047321.
vrt_full.coords["granule_id"] = ('acq_date', granule_da.data)
vrt_full.granule_id
<xarray.DataArray 'granule_id' (acq_date: 103)>
array(['S1A_IW_SLC__1SDV_20210502T121414_20210502T121441_037709_047321_900F',
'S1A_IW_SLC__1SDV_20210505T000307_20210505T000334_037745_047463_4DEF',
'S1A_IW_SLC__1SDV_20210509T120542_20210509T120609_037811_047676_9EBD',
'S1A_IW_SLC__1SDV_20210514T121349_20210514T121416_037884_047898_4702',
'S1A_IW_SLC__1SDV_20210514T121414_20210514T121442_037884_047898_FE6F',
'S1A_IW_SLC__1SDV_20210517T000308_20210517T000335_037920_0479A9_2A18',
'S1A_IW_SLC__1SDV_20210521T120543_20210521T120609_037986_047BBD_1CA0',
'S1A_IW_SLC__1SDV_20210526T121350_20210526T121417_038059_047DE5_0E67',
'S1A_IW_SLC__1SDV_20210529T000309_20210529T000336_038095_047EF4_5449',
'S1A_IW_SLC__1SDV_20210602T120543_20210602T120610_038161_0480FD_893F',
'S1A_IW_SLC__1SDV_20210607T121351_20210607T121418_038234_048318_9B64',
'S1A_IW_SLC__1SDV_20210610T000310_20210610T000337_038270_04841E_601A',
'S1A_IW_SLC__1SDV_20210614T120544_20210614T120611_038336_04862F_337B',
'S1A_IW_SLC__1SDV_20210619T121352_20210619T121419_038409_04884C_1DBA',
'S1A_IW_SLC__1SDV_20210622T000310_20210622T000337_038445_04895A_842E',
'S1A_IW_SLC__1SDV_20210626T120545_20210626T120612_038511_048B6C_9585',
'S1A_IW_SLC__1SDV_20210701T121352_20210701T121419_038584_048D87_F95C',
'S1A_IW_SLC__1SDV_20210701T121417_20210701T121445_038584_048D87_C79E',
'S1A_IW_SLC__1SDV_20210704T000311_20210704T000338_038620_048E99_1BAA',
'S1A_IW_SLC__1SDV_20210708T120545_20210708T120612_038686_0490AD_DD9E',
...
'S1A_IW_SLC__1SDV_20220317T120545_20220317T120612_042361_050CE5_4CCF',
'S1A_IW_SLC__1SDV_20220322T121353_20220322T121420_042434_050F59_A1B6',
'S1A_IW_SLC__1SDV_20220325T000312_20220325T000339_042470_051096_BA6F',
'S1A_IW_SLC__1SDV_20220329T120546_20220329T120613_042536_0512D7_425A',
'S1A_IW_SLC__1SDV_20220403T121418_20220403T121446_042609_05154A_51E7',
'S1A_IW_SLC__1SDV_20220403T121353_20220403T121420_042609_05154A_3FE7',
'S1A_IW_SLC__1SDV_20220406T000312_20220406T000339_042645_051682_4B40',
'S1A_IW_SLC__1SDV_20220410T120546_20220410T120613_042711_0518C0_3349',
'S1A_IW_SLC__1SDV_20220415T121353_20220415T121420_042784_051B30_EB87',
'S1A_IW_SLC__1SDV_20220418T000312_20220418T000339_042820_051C63_CAB2',
'S1A_IW_SLC__1SDV_20220422T120547_20220422T120614_042886_051E96_41EE',
'S1A_IW_SLC__1SDV_20220427T121354_20220427T121421_042959_0520EE_A42D',
'S1A_IW_SLC__1SDV_20220430T000313_20220430T000340_042995_05221E_46A1',
'S1A_IW_SLC__1SDV_20220504T120547_20220504T120614_043061_05245F_6F62',
'S1A_IW_SLC__1SDV_20220509T121355_20220509T121422_043134_0526C4_092E',
'S1A_IW_SLC__1SDV_20220516T120548_20220516T120615_043236_0529DD_42D2',
'S1A_IW_SLC__1SDV_20220521T121420_20220521T121448_043309_052C00_493D',
'S1A_IW_SLC__1SDV_20220521T121420_20220521T121448_043309_052C00_493D',
'S1A_IW_SLC__1SDV_20220521T121356_20220521T121423_043309_052C00_FE88'],
dtype='<U67')
Coordinates: (12/14)
* acq_date (acq_date) datetime64[ns] 2021-05-02T12...
spatial_ref int64 0
sensor (acq_date) <U3 'S1A' 'S1A' ... 'S1A' 'S1A'
beam_mode (acq_date) <U2 'IW' 'IW' ... 'IW' 'IW'
polarisation_type (acq_date) <U2 'DV' 'DV' ... 'DV' 'DV'
orbit_type (acq_date) <U1 '_' '_' '_' ... '_' '_' '_'
... ...
output_type (acq_date) <U1 'p' 'p' 'p' ... 'p' 'p' 'p'
masked (acq_date) <U1 'u' 'u' 'u' ... 'u' 'u' 'u'
filtered (acq_date) <U1 'n' 'n' 'n' ... 'n' 'n' 'n'
area (acq_date) <U1 'e' 'e' 'e' ... 'e' 'e' 'e'
product_id (acq_date) <U4 '748F' '0D1E' ... 'BD36'
granule_id (acq_date) <U67 'S1A_IW_SLC__1SDV_20210...Add orbital direction coord#
Next we want to add orbital direction as a coordinate to the xarray object. To do this we will:
Use
acq_date.dt.hourto segment acquisitions by orbital directionUse
xarray.where()to translate hour to “asc” or “desc”Assign coord for orbital pass
Check the dt.hour accessor of the vrt_full.acquisition_time data array to confirm that’s what we want:
vrt_full.isel(acq_date=0)
<xarray.Dataset>
Dimensions: (x: 290, y: 396)
Coordinates: (12/16)
* x (x) float64 6.194e+05 ... 6.281e+05
* y (y) float64 3.102e+06 ... 3.09e+06
acq_date datetime64[ns] 2021-05-02T12:14:14
spatial_ref int64 0
sensor <U3 'S1A'
beam_mode <U2 'IW'
... ...
output_type <U1 'p'
masked <U1 'u'
filtered <U1 'n'
area <U1 'e'
product_id <U4 '748F'
granule_id <U67 'S1A_IW_SLC__1SDV_20210502T121414_...
Data variables:
vv (y, x) float32 dask.array<chunksize=(396, 290), meta=np.ndarray>
vh (y, x) float32 dask.array<chunksize=(396, 290), meta=np.ndarray>
ls (y, x) uint8 dask.array<chunksize=(396, 290), meta=np.ndarray>vrt_full.acq_date.dt.hour
<xarray.DataArray 'hour' (acq_date: 103)>
array([12, 0, 12, 12, 12, 0, 12, 12, 0, 12, 12, 0, 12, 12, 0, 12, 12, 12,
0, 12, 12, 12, 12, 0, 12, 12, 0, 12, 12, 0, 12, 12, 0, 12, 12, 0,
12, 12, 0, 12, 12, 0, 12, 12, 0, 12, 12, 0, 12, 12, 0, 12, 12, 0,
12, 12, 0, 12, 12, 0, 12, 12, 0, 12, 12, 0, 12, 12, 0, 12, 12, 0,
12, 12, 0, 12, 12, 0, 12, 12, 0, 12, 12, 0, 12, 12, 0, 12, 12, 12,
0, 12, 12, 0, 12, 12, 0, 12, 12, 12, 12, 12, 12])
Coordinates: (12/14)
* acq_date (acq_date) datetime64[ns] 2021-05-02T12...
spatial_ref int64 0
sensor (acq_date) <U3 'S1A' 'S1A' ... 'S1A' 'S1A'
beam_mode (acq_date) <U2 'IW' 'IW' ... 'IW' 'IW'
polarisation_type (acq_date) <U2 'DV' 'DV' ... 'DV' 'DV'
orbit_type (acq_date) <U1 '_' '_' '_' ... '_' '_' '_'
... ...
output_type (acq_date) <U1 'p' 'p' 'p' ... 'p' 'p' 'p'
masked (acq_date) <U1 'u' 'u' 'u' ... 'u' 'u' 'u'
filtered (acq_date) <U1 'n' 'n' 'n' ... 'n' 'n' 'n'
area (acq_date) <U1 'e' 'e' 'e' ... 'e' 'e' 'e'
product_id (acq_date) <U4 '748F' '0D1E' ... 'BD36'
granule_id (acq_date) <U67 'S1A_IW_SLC__1SDV_20210...Assign the above object as a coordinate of vrt_full:
vrt_full.coords['orbital_dir'] = ('acq_date', xr.where(vrt_full.acq_date.dt.hour.data== 0, 'asc','desc'))
vrt_full
<xarray.Dataset>
Dimensions: (x: 290, y: 396, acq_date: 103)
Coordinates: (12/17)
* x (x) float64 6.194e+05 ... 6.281e+05
* y (y) float64 3.102e+06 ... 3.09e+06
* acq_date (acq_date) datetime64[ns] 2021-05-02T12...
spatial_ref int64 0
sensor (acq_date) <U3 'S1A' 'S1A' ... 'S1A' 'S1A'
beam_mode (acq_date) <U2 'IW' 'IW' ... 'IW' 'IW'
... ...
masked (acq_date) <U1 'u' 'u' 'u' ... 'u' 'u' 'u'
filtered (acq_date) <U1 'n' 'n' 'n' ... 'n' 'n' 'n'
area (acq_date) <U1 'e' 'e' 'e' ... 'e' 'e' 'e'
product_id (acq_date) <U4 '748F' '0D1E' ... 'BD36'
granule_id (acq_date) <U67 'S1A_IW_SLC__1SDV_20210...
orbital_dir (acq_date) <U4 'desc' 'asc' ... 'desc'
Data variables:
vv (acq_date, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
vh (acq_date, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
ls (acq_date, y, x) uint8 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>Handle nodata values#
Another important difference between the xr.open_mfdataset() and VRT approaches is the handling of the nodata values. In the object built using xr.open_mfdataset(), nodata values are set to NaN while building the object from VRT files converts nodata values to zeros. This can cause a stream of problems later on so it is best to convert them to NaNs now. You can check the handling of nodata values in your dataset by using the rio.nodata accessor (shown below) which you can read about here.
vrt_full.vv.isel(acq_date=0).rio.nodata
0.0
^ We want this to be a nan.
So using xr.Dataset.where() (don’t think this will actually overwrite the official nodata value, but it should replace the zeros:
vrt_full = vrt_full.where(vrt_full.vv != 0., np.nan, drop=False)
vrt_full
<xarray.Dataset>
Dimensions: (acq_date: 103, y: 396, x: 290)
Coordinates: (12/17)
* x (x) float64 6.194e+05 ... 6.281e+05
* y (y) float64 3.102e+06 ... 3.09e+06
* acq_date (acq_date) datetime64[ns] 2021-05-02T12...
spatial_ref int64 0
sensor (acq_date) <U3 'S1A' 'S1A' ... 'S1A' 'S1A'
beam_mode (acq_date) <U2 'IW' 'IW' ... 'IW' 'IW'
... ...
masked (acq_date) <U1 'u' 'u' 'u' ... 'u' 'u' 'u'
filtered (acq_date) <U1 'n' 'n' 'n' ... 'n' 'n' 'n'
area (acq_date) <U1 'e' 'e' 'e' ... 'e' 'e' 'e'
product_id (acq_date) <U4 '748F' '0D1E' ... 'BD36'
granule_id (acq_date) <U67 'S1A_IW_SLC__1SDV_20210...
orbital_dir (acq_date) <U4 'desc' 'asc' ... 'desc'
Data variables:
vv (acq_date, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
vh (acq_date, y, x) float32 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>
ls (acq_date, y, x) float64 dask.array<chunksize=(1, 396, 290), meta=np.ndarray>Quick visualization#
Before we move on to the next notebook, let’s take a quick look at the dataset. We will use the function defined earlier to convert the data from power scale to dB scale. This makes it easier to visualize variability in backscatter. Read more about SAR data scales here.
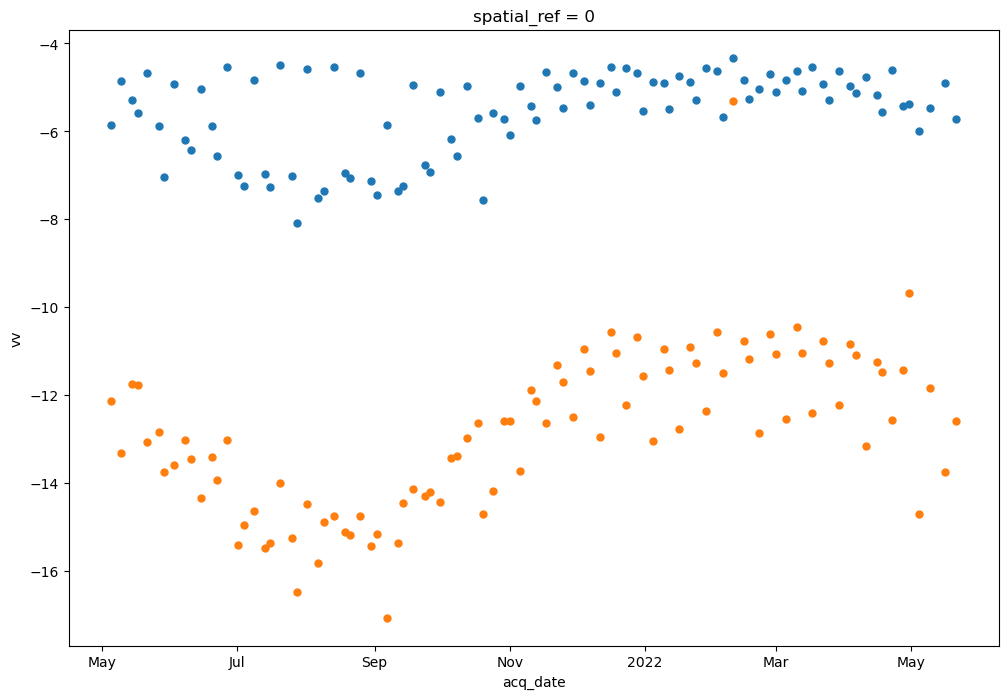
First let’s visualize backscatter variability over time. To do this, we will perform a reduction along the x and y dimensions of the dataset. We can use built-in xarray plotting methods to plot the 2-dimensional data (backscatter and time, after reducing along x and y)
(See the Xarray User Guide for more on visualizing Xarray objects)
fig, ax = plt.subplots(figsize=(12,8));
power_to_db(vrt_full.vh.mean(dim=['x','y'])).plot(ax=ax, marker='o', linestyle='None', markersize=5)
power_to_db(vrt_full.vv.mean(dim=['x','y'])).plot(ax=ax, marker='o', linestyle='None', markersize=5);
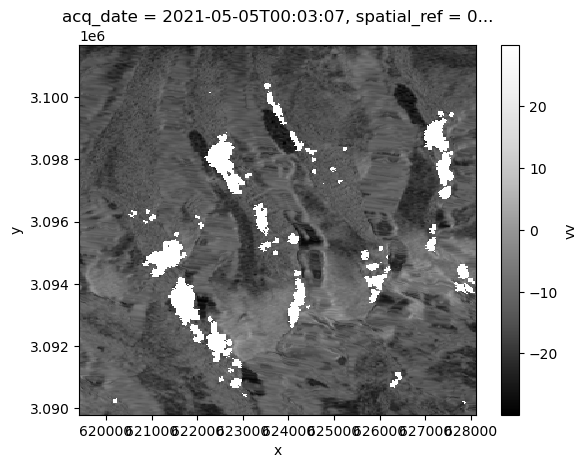
Now what if we wanted to take a snap-shot look of backscatter at a single time step? We can still use xarray plotting methods, but this time will pass a 3-dimensional object (x,y and backscatter). We need to select a single element along the time dimension on which to call the plotting method:
power_to_db(vrt_full.vv.isel(acq_date=1)).plot(cmap=plt.cm.Greys_r);
Now we have all of the same metadata attached to our xarray datacube of Sentinel-1 RTC imagery that we had using the xr.open_mfdataset() approach, but we’ve done this with much less computational expense and a chunking strategy that is more appropriate for the nature of the dataset.
We’ll store this object to use it in a later notebook.
%store vrt_full
Stored 'vrt_full' (Dataset)
Next#
The next notebook on preliminary dataset inspection demonstrates how to explore and familiarize oneself with the dataset now that we have it read in and organized.